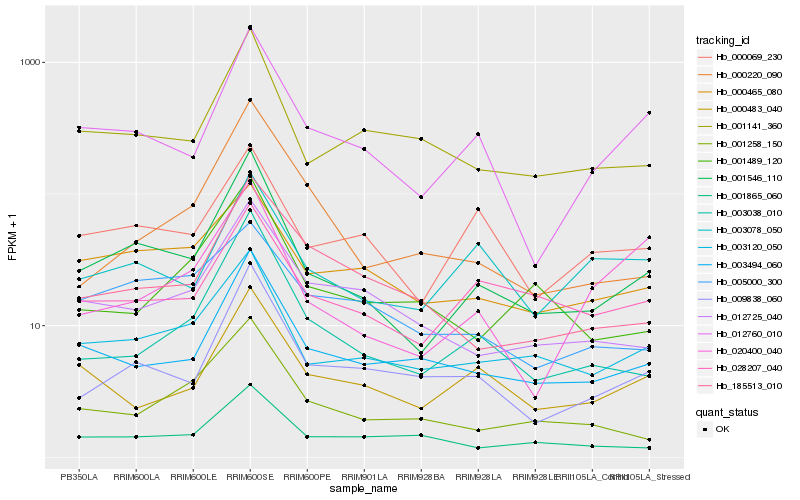
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001546_110 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 3 [Jatropha curcas] |
| 2 |
Hb_003120_050 |
0.1356752455 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 3 |
Hb_012760_010 |
0.1406115274 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 4 |
Hb_009838_060 |
0.1480563201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003494_060 |
0.1492355629 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003038_010 |
0.1506376746 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_001141_360 |
0.1626270483 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 8 |
Hb_000465_080 |
0.1702506302 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 9 |
Hb_028207_040 |
0.1706142854 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 10 |
Hb_000069_230 |
0.173840461 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 11 |
Hb_000483_040 |
0.1759846306 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 12 |
Hb_012725_040 |
0.1767381313 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 13 |
Hb_001258_150 |
0.1778561458 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 14 |
Hb_003078_050 |
0.1831459174 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 15 |
Hb_001865_060 |
0.1902179031 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 16 |
Hb_185513_010 |
0.1908484898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001489_120 |
0.1926812266 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000220_090 |
0.1929401755 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 19 |
Hb_020400_040 |
0.1970722505 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 20 |
Hb_005000_300 |
0.1974296814 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |