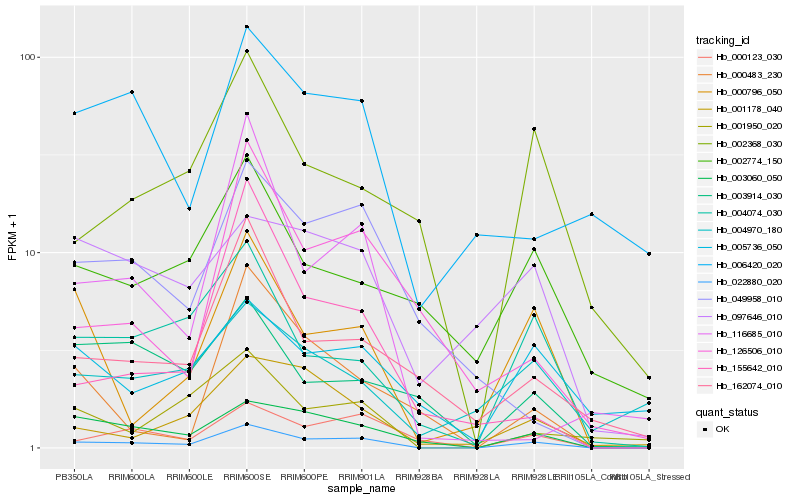
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022880_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 2 |
Hb_003060_050 |
0.1975927852 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 3 |
Hb_000123_030 |
0.2034710113 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_004074_030 |
0.2150647442 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000796_050 |
0.2229954971 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 6 |
Hb_001178_040 |
0.2354963729 |
desease resistance |
Gene Name: ABC_trans_N |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 7 |
Hb_001950_020 |
0.2378636354 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 8 |
Hb_162074_010 |
0.2428332643 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 9 |
Hb_002368_030 |
0.2560563366 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 10 |
Hb_049958_010 |
0.2596718037 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 11 |
Hb_116685_010 |
0.2607810906 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 12 |
Hb_097646_010 |
0.2623409889 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 13 |
Hb_003914_030 |
0.2646500459 |
- |
- |
- |
| 14 |
Hb_126506_010 |
0.2659671501 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 15 |
Hb_005736_050 |
0.2664134041 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 16 |
Hb_002774_150 |
0.2664477706 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 17 |
Hb_155642_010 |
0.2672730143 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 18 |
Hb_006420_020 |
0.2682143381 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 19 |
Hb_000483_230 |
0.2707201202 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_004970_180 |
0.2716565095 |
- |
- |
- |