| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
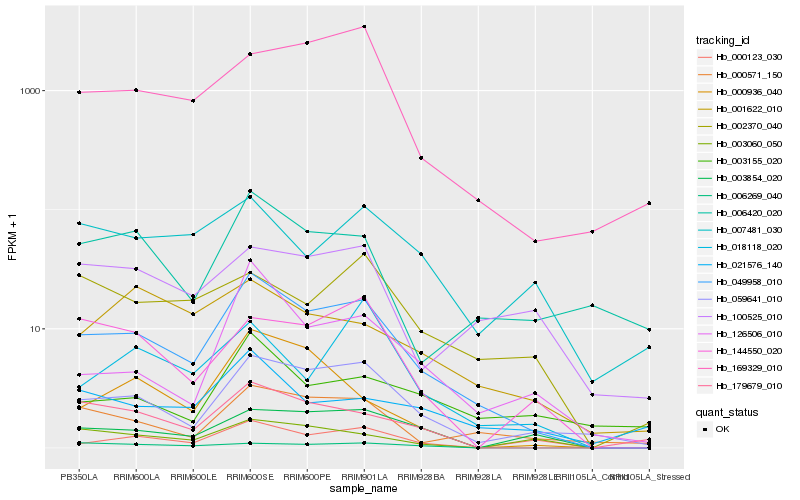
Hb_049958_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 2 |
Hb_059641_010 |
0.1531529181 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 3 |
Hb_000571_150 |
0.1960954303 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_179679_010 |
0.2000846969 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Fragaria vesca subsp. vesca] |
| 5 |
Hb_126506_010 |
0.2062860645 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 6 |
Hb_169329_010 |
0.2071737513 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000123_030 |
0.2166354474 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 8 |
Hb_144550_020 |
0.2187793029 |
- |
- |
- |
| 9 |
Hb_003854_020 |
0.21971268 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_006420_020 |
0.2253397488 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_006269_040 |
0.2282131327 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000936_040 |
0.2282563968 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 13 |
Hb_021576_140 |
0.2307414642 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 14 |
Hb_002370_040 |
0.231262097 |
- |
- |
PREDICTED: uncharacterized protein LOC105644336 [Jatropha curcas] |
| 15 |
Hb_001622_010 |
0.2334162927 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_003060_050 |
0.2336593468 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 17 |
Hb_018118_020 |
0.2348258494 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_100525_010 |
0.2356574196 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 19 |
Hb_007481_030 |
0.2374016224 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |
| 20 |
Hb_003155_020 |
0.2376319867 |
- |
- |
Rpp4C3 [Medicago truncatula] |