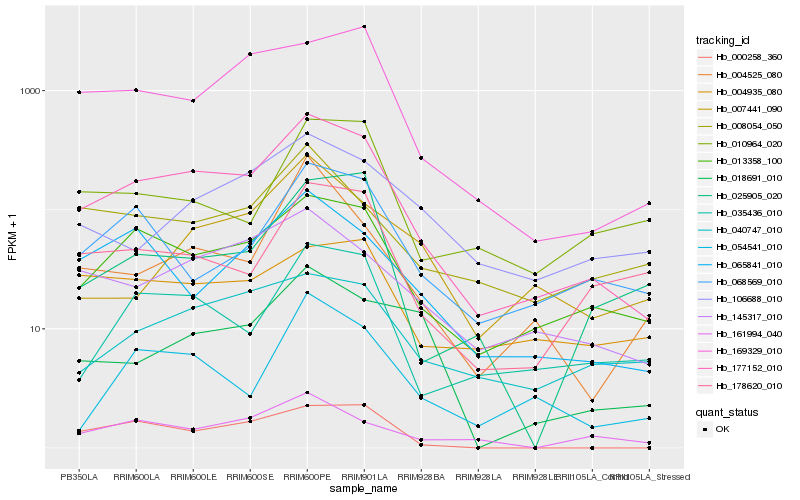
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_177152_010 |
0.0 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 2 |
Hb_013358_100 |
0.1264568874 |
- |
- |
unknown [Medicago truncatula] |
| 3 |
Hb_145317_010 |
0.1663011061 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 4 |
Hb_169329_010 |
0.1739169479 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 5 |
Hb_035436_010 |
0.176103837 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 6 |
Hb_065841_010 |
0.1765645154 |
- |
- |
hypothetical protein JCGZ_18376 [Jatropha curcas] |
| 7 |
Hb_178620_010 |
0.1859044027 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |
| 8 |
Hb_040747_010 |
0.1871935662 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 9 |
Hb_000258_360 |
0.1880124854 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 10 |
Hb_068569_010 |
0.1885467706 |
- |
- |
- |
| 11 |
Hb_008054_050 |
0.1920853716 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 12 |
Hb_054541_010 |
0.1951465171 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 13 |
Hb_004525_080 |
0.1960650353 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 14 |
Hb_106688_010 |
0.1963292693 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 15 |
Hb_161994_040 |
0.1972130241 |
- |
- |
- |
| 16 |
Hb_004935_080 |
0.1992785597 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 17 |
Hb_018691_010 |
0.205426816 |
- |
- |
- |
| 18 |
Hb_025905_020 |
0.2056459669 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 19 |
Hb_010964_020 |
0.2072419076 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 20 |
Hb_007441_090 |
0.2082345708 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |