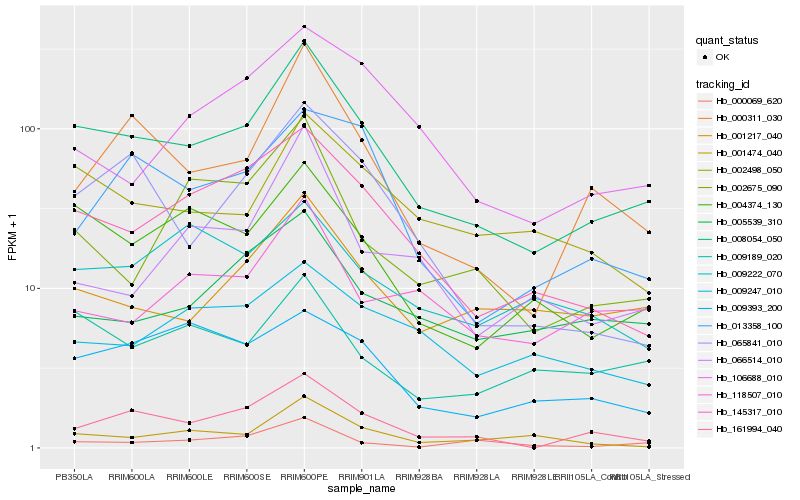
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008054_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 2 |
Hb_145317_010 |
0.1353604692 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 3 |
Hb_004374_130 |
0.1381849797 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 4 |
Hb_161994_040 |
0.1493862969 |
- |
- |
- |
| 5 |
Hb_000311_030 |
0.1522719827 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 6 |
Hb_002498_050 |
0.1571234582 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 7 |
Hb_001217_040 |
0.1611761986 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 8 |
Hb_065841_010 |
0.1654384062 |
- |
- |
hypothetical protein JCGZ_18376 [Jatropha curcas] |
| 9 |
Hb_009189_020 |
0.1694279218 |
- |
- |
PREDICTED: CASP-like protein 4B4 [Jatropha curcas] |
| 10 |
Hb_005539_310 |
0.1750841658 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001474_040 |
0.1778043718 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 12 |
Hb_002675_090 |
0.1794066992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_118507_010 |
0.1844059808 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 14 |
Hb_106688_010 |
0.1844901716 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 15 |
Hb_013358_100 |
0.1851298374 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_009247_010 |
0.1853311478 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_009393_200 |
0.1859855381 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 18 |
Hb_066514_010 |
0.1888051288 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 19 |
Hb_000069_620 |
0.1890428746 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_009222_070 |
0.1904644509 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |