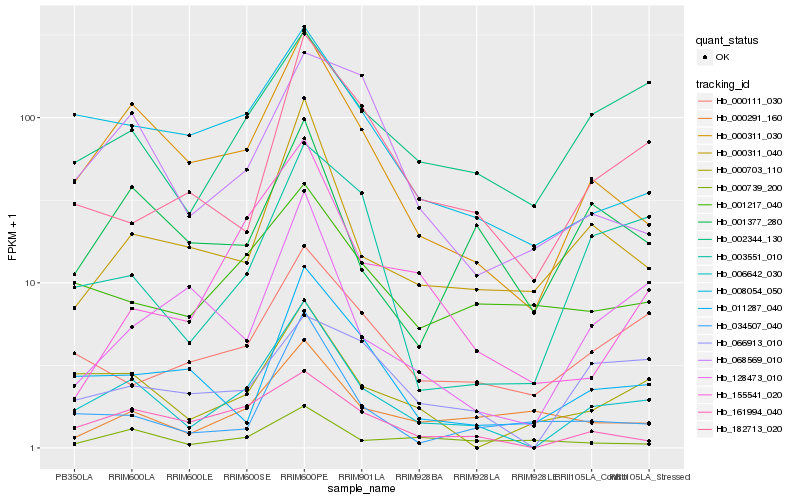
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006642_030 |
0.0 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 2 |
Hb_000311_030 |
0.1493174131 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 3 |
Hb_161994_040 |
0.1888412207 |
- |
- |
- |
| 4 |
Hb_000703_110 |
0.2020127508 |
- |
- |
- |
| 5 |
Hb_000311_040 |
0.2027699574 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 6 |
Hb_008054_050 |
0.2061830866 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 7 |
Hb_002344_130 |
0.216814472 |
- |
- |
PREDICTED: copper transporter 5 [Jatropha curcas] |
| 8 |
Hb_000111_030 |
0.2256516394 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 9 |
Hb_003551_010 |
0.2264995176 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 10 |
Hb_011287_040 |
0.2283185304 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_182713_020 |
0.2288498591 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 12 |
Hb_034507_040 |
0.2290196434 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 13 |
Hb_001377_280 |
0.2313373532 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 14 |
Hb_001217_040 |
0.2323277249 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 15 |
Hb_066913_010 |
0.2360515231 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 16 |
Hb_128473_010 |
0.2370118517 |
- |
- |
- |
| 17 |
Hb_155541_020 |
0.2374515477 |
- |
- |
- |
| 18 |
Hb_000739_200 |
0.2374767905 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 19 |
Hb_000291_160 |
0.2387206028 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 20 |
Hb_068569_010 |
0.2413678035 |
- |
- |
- |