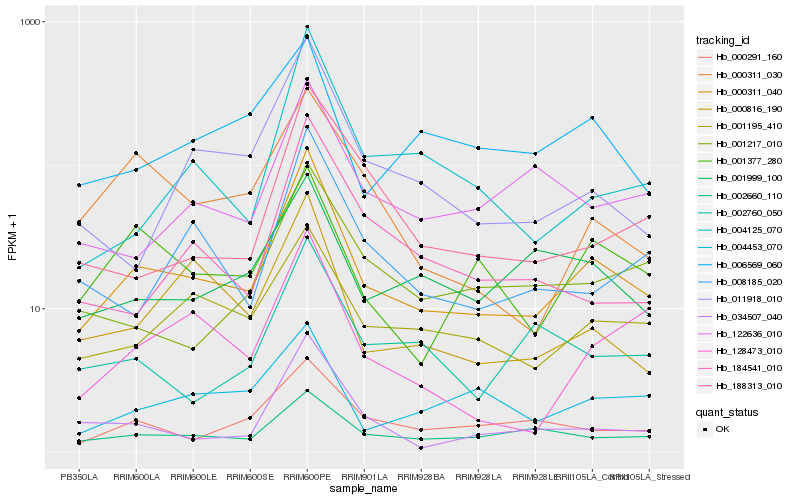
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000311_040 |
0.0 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 2 |
Hb_001195_410 |
0.1662100924 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 3 |
Hb_034507_040 |
0.1744076606 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 4 |
Hb_000816_190 |
0.1765002389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000291_160 |
0.1816084189 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 6 |
Hb_011918_010 |
0.181650119 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 7 |
Hb_008185_020 |
0.1816892385 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 8 |
Hb_001377_280 |
0.1832960786 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 9 |
Hb_002660_110 |
0.1836844878 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004125_070 |
0.1837372635 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 11 |
Hb_128473_010 |
0.18958802 |
- |
- |
- |
| 12 |
Hb_122636_010 |
0.1906630056 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 13 |
Hb_001999_100 |
0.1911646174 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 14 |
Hb_001217_010 |
0.1916824494 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 15 |
Hb_004453_070 |
0.1940640552 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000311_030 |
0.1948846915 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 17 |
Hb_002760_050 |
0.1949063219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006569_060 |
0.1960471264 |
- |
- |
plasma membrane intrinsic protein PIP2;3 [Hevea brasiliensis] |
| 19 |
Hb_184541_010 |
0.1967982144 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 20 |
Hb_188313_010 |
0.1982548682 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |