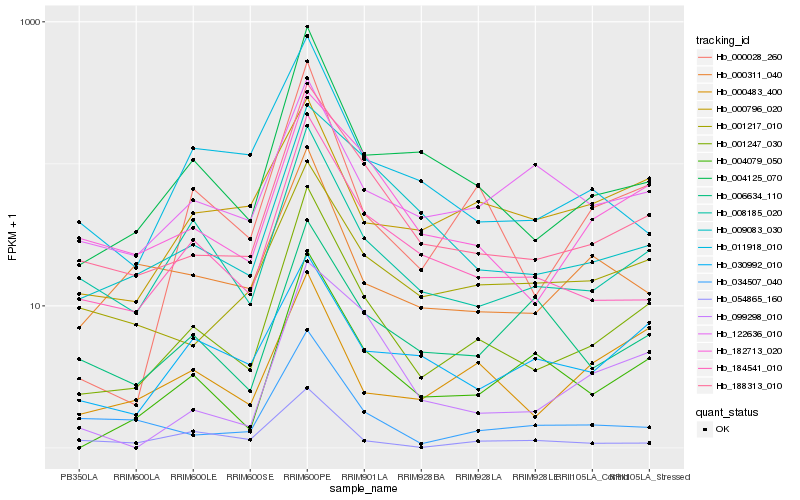
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_188313_010 |
0.1250216313 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 3 |
Hb_004125_070 |
0.1386128815 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 4 |
Hb_000028_260 |
0.1434300527 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 5 |
Hb_008185_020 |
0.1458264474 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 6 |
Hb_184541_010 |
0.1587501062 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 7 |
Hb_004079_050 |
0.1602654852 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 8 |
Hb_182713_020 |
0.171806688 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 9 |
Hb_001217_010 |
0.1969452889 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 10 |
Hb_034507_040 |
0.1989387953 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 11 |
Hb_099298_010 |
0.2001352253 |
- |
- |
- |
| 12 |
Hb_011918_010 |
0.2006206782 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 13 |
Hb_000311_040 |
0.2028334011 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 14 |
Hb_000483_400 |
0.2045683854 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_009083_030 |
0.2050956954 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 16 |
Hb_030992_010 |
0.2082589385 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 17 |
Hb_122636_010 |
0.2105195921 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 18 |
Hb_054865_160 |
0.211352235 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 19 |
Hb_000796_020 |
0.2123679267 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 20 |
Hb_006634_110 |
0.2136504627 |
- |
- |
BnaC04g40780D [Brassica napus] |