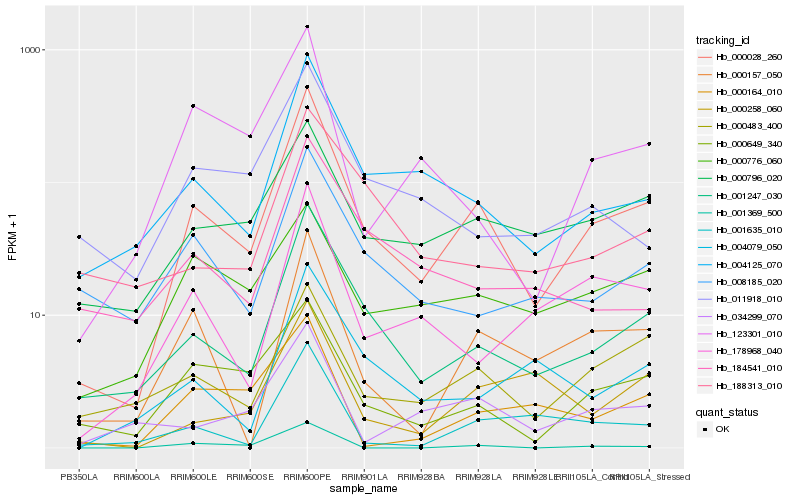
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_260 |
0.0 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 2 |
Hb_001247_030 |
0.1434300527 |
- |
- |
- |
| 3 |
Hb_004125_070 |
0.1839524177 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 4 |
Hb_000649_340 |
0.1990752275 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_000483_400 |
0.2031463069 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000157_050 |
0.2033067755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000796_020 |
0.2104435293 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 8 |
Hb_178968_040 |
0.2127093807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_123301_010 |
0.2211778014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008185_020 |
0.2237098117 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 11 |
Hb_004079_050 |
0.2276134871 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 12 |
Hb_034299_070 |
0.2286883896 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 13 |
Hb_001635_010 |
0.2303913756 |
- |
- |
- |
| 14 |
Hb_011918_010 |
0.2311881739 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 15 |
Hb_000258_060 |
0.2371703428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_184541_010 |
0.2384523154 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 17 |
Hb_000164_010 |
0.2388577035 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 18 |
Hb_188313_010 |
0.2397823633 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 19 |
Hb_000776_060 |
0.2423186795 |
- |
- |
histone H4 [Zea mays] |
| 20 |
Hb_001369_500 |
0.2462791032 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |