| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
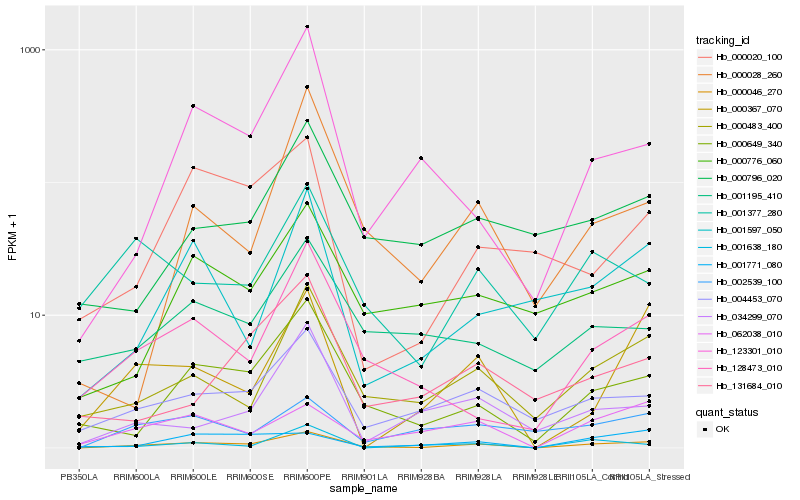
Hb_000046_270 |
0.0 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000649_340 |
0.1992354414 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 3 |
Hb_000483_400 |
0.2115737254 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_004453_070 |
0.214546886 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_128473_010 |
0.2300828539 |
- |
- |
- |
| 6 |
Hb_062038_010 |
0.2401699215 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC104882266 isoform X2 [Vitis vinifera] |
| 7 |
Hb_000776_060 |
0.2421647114 |
- |
- |
histone H4 [Zea mays] |
| 8 |
Hb_000367_070 |
0.2551974466 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 9 |
Hb_001597_050 |
0.255369131 |
- |
- |
- |
| 10 |
Hb_001377_280 |
0.2566189508 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 11 |
Hb_123301_010 |
0.2570766611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001195_410 |
0.2594575283 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 13 |
Hb_000796_020 |
0.2600533794 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 14 |
Hb_002539_100 |
0.2608529537 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 15 |
Hb_001771_080 |
0.2626456454 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Amborella trichopoda] |
| 16 |
Hb_000028_260 |
0.2647840718 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 17 |
Hb_001638_180 |
0.2654383264 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 18 |
Hb_000020_100 |
0.2688847351 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 19 |
Hb_034299_070 |
0.2694996176 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 20 |
Hb_131684_010 |
0.2724211969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |