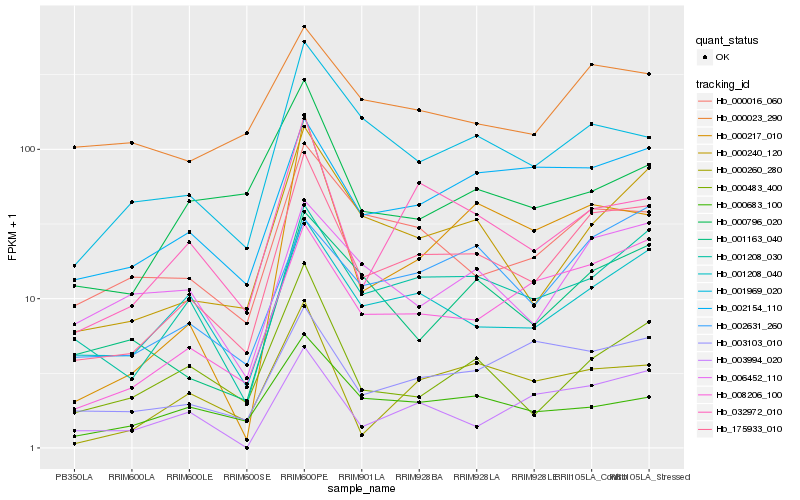
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_175933_010 |
0.0 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 2 |
Hb_000240_120 |
0.1347570754 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 3 |
Hb_001208_030 |
0.1382661962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008206_100 |
0.1394955263 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 5 |
Hb_032972_010 |
0.1441056536 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 6 |
Hb_001208_040 |
0.1524269499 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 7 |
Hb_001969_020 |
0.1570142874 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 8 |
Hb_000016_060 |
0.1575620831 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 9 |
Hb_002154_110 |
0.1619878482 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 10 |
Hb_000260_280 |
0.1675011304 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 11 |
Hb_001163_040 |
0.1679326043 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 12 |
Hb_000483_400 |
0.1723909635 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_003103_010 |
0.1747403291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003994_020 |
0.1792617918 |
- |
- |
- |
| 15 |
Hb_002631_260 |
0.1804171181 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 16 |
Hb_000217_010 |
0.1815544853 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 17 |
Hb_006452_110 |
0.1819202231 |
- |
- |
Ubiquinol-cytochrome c reductase complex 7.8 kDa protein, putative [Ricinus communis] |
| 18 |
Hb_000023_290 |
0.1839287797 |
- |
- |
steroid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000683_100 |
0.1845013999 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 20 |
Hb_000796_020 |
0.1848476542 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |