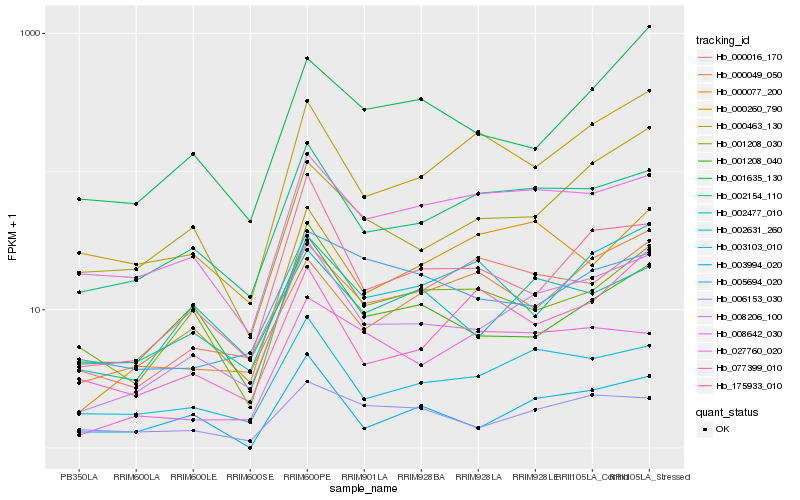
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008206_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 2 |
Hb_002154_110 |
0.1211269944 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 3 |
Hb_003103_010 |
0.1220250078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_175933_010 |
0.1394955263 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 5 |
Hb_000260_790 |
0.1540450992 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 12 isoform 1 [Theobroma cacao] |
| 6 |
Hb_003994_020 |
0.1550500291 |
- |
- |
- |
| 7 |
Hb_008642_030 |
0.1556345263 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001208_030 |
0.1579321205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002477_010 |
0.1634400492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002631_260 |
0.1679790329 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 11 |
Hb_006153_030 |
0.1718437535 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 12 |
Hb_000016_170 |
0.1724329945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000463_130 |
0.1726590454 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000077_200 |
0.1749247522 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 15 |
Hb_001208_040 |
0.1766074797 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 16 |
Hb_001635_130 |
0.1781742739 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 17 |
Hb_000049_050 |
0.1822279278 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 18 |
Hb_077399_010 |
0.1823256696 |
- |
- |
hypothetical protein POPTR_0003s09910g [Populus trichocarpa] |
| 19 |
Hb_027760_020 |
0.1828181022 |
- |
- |
hypothetical protein M569_04462, partial [Genlisea aurea] |
| 20 |
Hb_005694_020 |
0.1836797981 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |