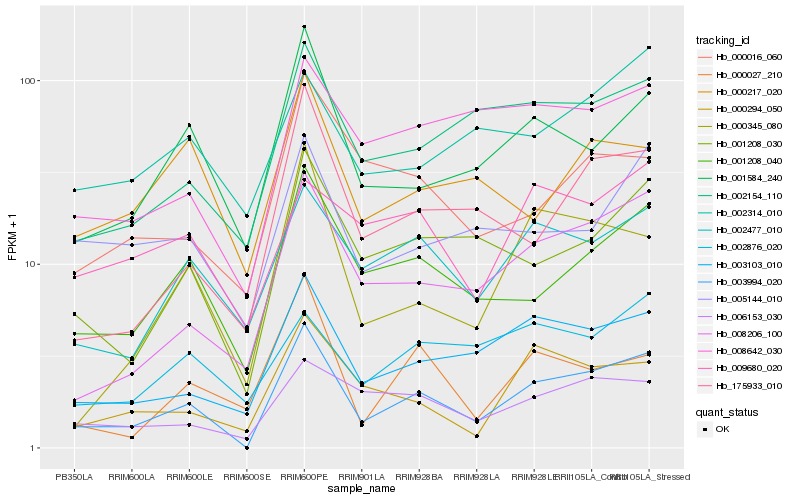
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_008206_100 |
0.1550500291 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 3 |
Hb_001208_040 |
0.1610670724 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 4 |
Hb_003103_010 |
0.1612468525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001208_030 |
0.168343326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001584_240 |
0.1749281841 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 7 |
Hb_175933_010 |
0.1792617918 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 8 |
Hb_002477_010 |
0.1808723272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000345_080 |
0.1819258975 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 10 |
Hb_002154_110 |
0.1830357976 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 11 |
Hb_000294_050 |
0.1929185232 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 12 |
Hb_006153_030 |
0.1944292509 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 13 |
Hb_008642_030 |
0.1951596782 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005144_010 |
0.2035416461 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 15 |
Hb_000027_210 |
0.2050461199 |
- |
- |
Os01g0689300, partial [Oryza sativa Japonica Group] |
| 16 |
Hb_000016_060 |
0.2071252507 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 17 |
Hb_002876_020 |
0.2161027863 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 18 |
Hb_009680_020 |
0.2174267353 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 19 |
Hb_002314_010 |
0.2179748826 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 20 |
Hb_000217_020 |
0.2179992498 |
- |
- |
AMME syndrome candidateprotein 1 protein, putative [Ricinus communis] |