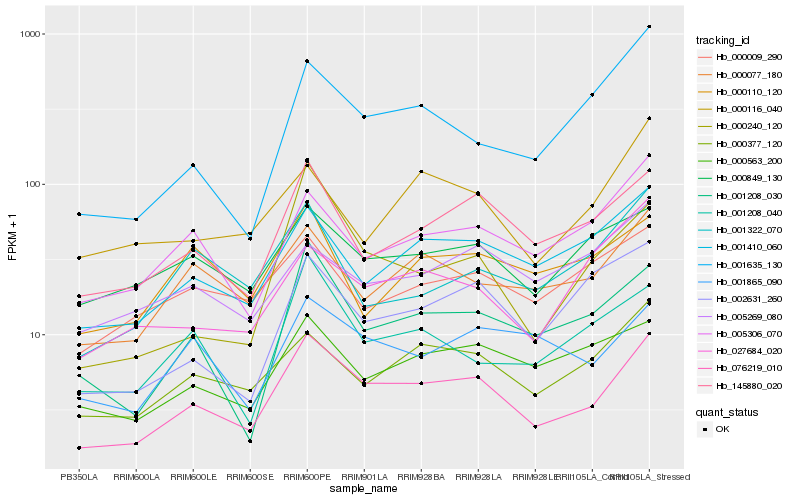
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_076219_010 |
0.0 |
- |
- |
PREDICTED: RNA pseudouridine synthase 4, mitochondrial [Jatropha curcas] |
| 2 |
Hb_145880_020 |
0.1213967114 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 3 |
Hb_001410_060 |
0.1324345375 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 4 |
Hb_000377_120 |
0.1330808592 |
- |
- |
PREDICTED: acyl carrier protein 3, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_000077_180 |
0.1369166482 |
- |
- |
- |
| 6 |
Hb_001208_030 |
0.1370801996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005306_070 |
0.1423710675 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 8 |
Hb_000849_130 |
0.14601962 |
- |
- |
PREDICTED: adrenodoxin-like protein, mitochondrial [Nelumbo nucifera] |
| 9 |
Hb_000563_200 |
0.1471280235 |
- |
- |
hypothetical protein JCGZ_18930 [Jatropha curcas] |
| 10 |
Hb_001865_090 |
0.1496688818 |
- |
- |
hypothetical protein VITISV_005773 [Vitis vinifera] |
| 11 |
Hb_002631_260 |
0.1536978864 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 12 |
Hb_001208_040 |
0.1543556467 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 13 |
Hb_000009_290 |
0.1566819433 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 14 |
Hb_001635_130 |
0.1626223623 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 15 |
Hb_001322_070 |
0.1642826957 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_005269_080 |
0.1648249739 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_000110_120 |
0.1649979021 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 18 |
Hb_000240_120 |
0.1654109783 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 19 |
Hb_027684_020 |
0.1659209418 |
- |
- |
ribosomal protein S10 (mitochondrion) [Nicotiana tabacum] |
| 20 |
Hb_000116_040 |
0.1663017566 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |