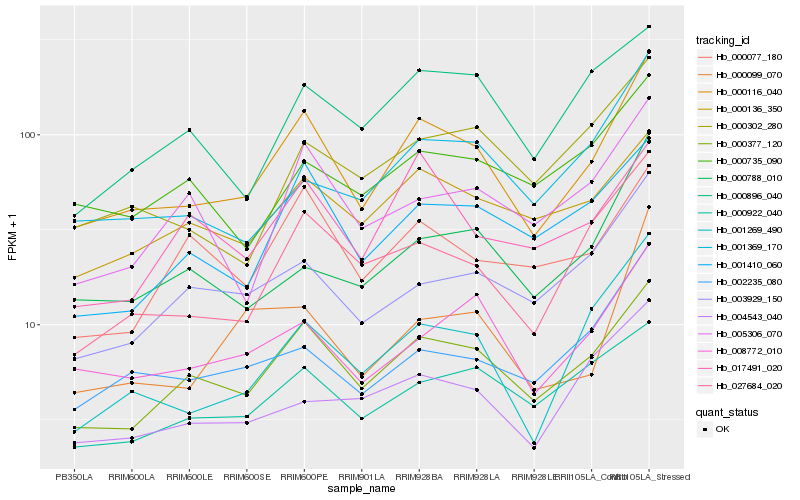
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_040 |
0.0 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000377_120 |
0.0849576931 |
- |
- |
PREDICTED: acyl carrier protein 3, mitochondrial isoform X2 [Jatropha curcas] |
| 3 |
Hb_027684_020 |
0.1097796178 |
- |
- |
ribosomal protein S10 (mitochondrion) [Nicotiana tabacum] |
| 4 |
Hb_001269_490 |
0.1174505689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008772_010 |
0.1201927308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001410_060 |
0.1245495075 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 7 |
Hb_000136_350 |
0.1282412072 |
- |
- |
PREDICTED: 40S ribosomal protein S5 [Nomascus leucogenys] |
| 8 |
Hb_002235_080 |
0.130541645 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 9 |
Hb_001369_170 |
0.1322998667 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1-like [Gossypium raimondii] |
| 10 |
Hb_003929_150 |
0.1333745764 |
- |
- |
PREDICTED: rRNA-processing protein fcf2-like [Jatropha curcas] |
| 11 |
Hb_000922_040 |
0.1355060298 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 12 |
Hb_000788_010 |
0.1362546887 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 13 |
Hb_000302_280 |
0.1384653817 |
- |
- |
hypothetical protein ZEAMMB73_772347 [Zea mays] |
| 14 |
Hb_000896_040 |
0.1392235749 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 15 |
Hb_005306_070 |
0.1401418229 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 16 |
Hb_000077_180 |
0.1403182378 |
- |
- |
- |
| 17 |
Hb_004543_040 |
0.1413939302 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000099_070 |
0.1416639706 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 19 |
Hb_017491_020 |
0.1420447553 |
- |
- |
hypothetical protein B456_011G056700 [Gossypium raimondii] |
| 20 |
Hb_000735_090 |
0.1433241449 |
- |
- |
PREDICTED: 40S ribosomal protein S8-like [Gossypium raimondii] |