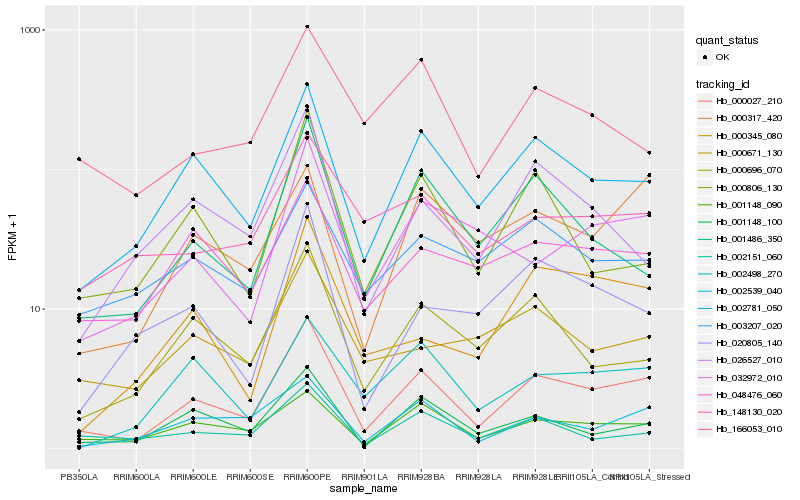
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_210 |
0.0 |
- |
- |
Os01g0689300, partial [Oryza sativa Japonica Group] |
| 2 |
Hb_002781_050 |
0.1317709272 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 3 |
Hb_001148_090 |
0.1559077134 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 4 |
Hb_001148_100 |
0.1676381299 |
- |
- |
- |
| 5 |
Hb_002539_040 |
0.1681657937 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_002151_060 |
0.1762210068 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc4 [Jatropha curcas] |
| 7 |
Hb_001486_350 |
0.1771153693 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_032972_010 |
0.1771184424 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 9 |
Hb_000696_070 |
0.1781566044 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 10 |
Hb_048476_060 |
0.1791373668 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_002498_270 |
0.1860203213 |
- |
- |
prephenate dehydratase, putative [Ricinus communis] |
| 12 |
Hb_000806_130 |
0.1891426063 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 13 |
Hb_000317_420 |
0.1896074391 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 14 |
Hb_000345_080 |
0.1915241533 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 15 |
Hb_148130_020 |
0.1926079936 |
- |
- |
- |
| 16 |
Hb_003207_020 |
0.1946628303 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 17 |
Hb_000671_130 |
0.1946925688 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_026527_010 |
0.1948145697 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 19 |
Hb_166053_010 |
0.1958243343 |
- |
- |
glyceraldehyde-3-phosphate dehydrogenase, partial [Vernicia fordii] |
| 20 |
Hb_020805_140 |
0.196194933 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |