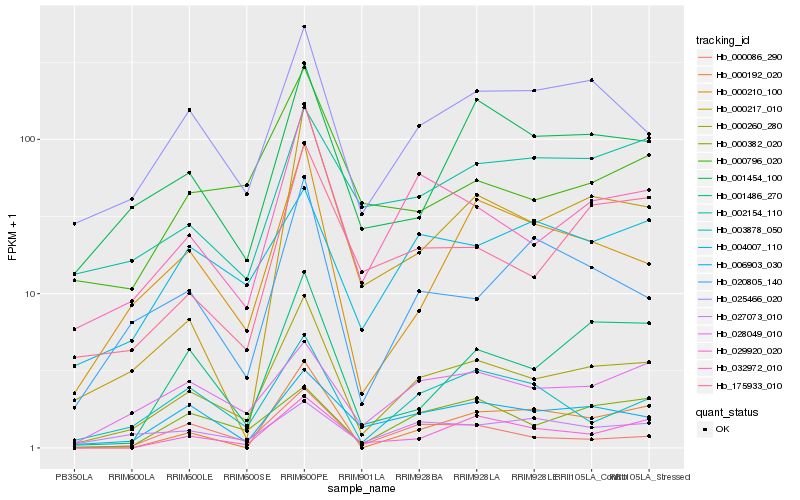
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_280 |
0.0 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 2 |
Hb_032972_010 |
0.1279517492 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 3 |
Hb_025466_020 |
0.1460578615 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 4 |
Hb_000210_100 |
0.1606223091 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 5 |
Hb_001454_100 |
0.164815216 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 6 |
Hb_029920_020 |
0.1661077846 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 7 |
Hb_006903_030 |
0.1664364143 |
- |
- |
PREDICTED: uncharacterized protein LOC105640666 [Jatropha curcas] |
| 8 |
Hb_175933_010 |
0.1675011304 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 9 |
Hb_001486_270 |
0.1724590369 |
- |
- |
Ann10 [Manihot esculenta] |
| 10 |
Hb_028049_010 |
0.1733751994 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_020805_140 |
0.1754560376 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 12 |
Hb_000217_010 |
0.1767078574 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 13 |
Hb_003878_050 |
0.1789508732 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 14 |
Hb_000382_020 |
0.1819299994 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 15 |
Hb_027073_010 |
0.1860080565 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 16 |
Hb_000192_020 |
0.1867904029 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 17 |
Hb_004007_110 |
0.1879539314 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 18 |
Hb_000796_020 |
0.1903889951 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 19 |
Hb_000086_290 |
0.1917901987 |
- |
- |
PREDICTED: TBC1 domain family member 15-like [Jatropha curcas] |
| 20 |
Hb_002154_110 |
0.1918118812 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |