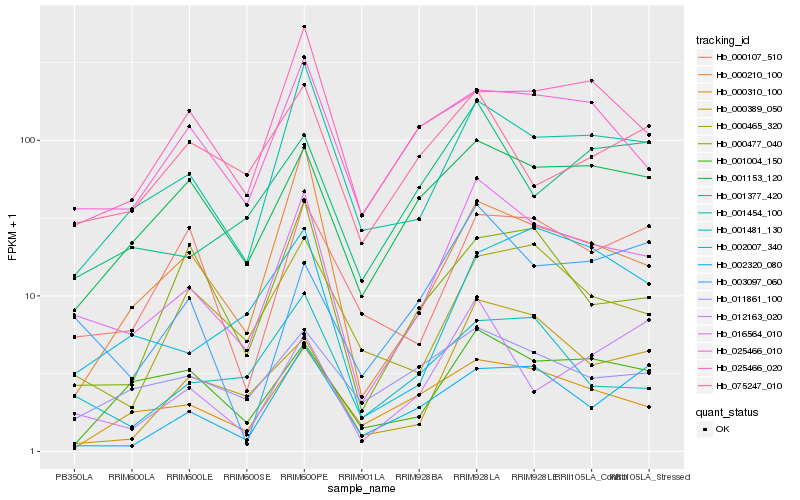
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016564_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101764501 [Setaria italica] |
| 2 |
Hb_001454_100 |
0.1562460618 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 3 |
Hb_001153_120 |
0.1822235697 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 4 |
Hb_000389_050 |
0.1868542714 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 5 |
Hb_001004_150 |
0.1908403971 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 6 |
Hb_025466_010 |
0.1924534195 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003097_060 |
0.1953508096 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 8 |
Hb_000465_320 |
0.1955849668 |
- |
- |
PREDICTED: nudix hydrolase 8 [Jatropha curcas] |
| 9 |
Hb_000310_100 |
0.1957241513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000210_100 |
0.1964969742 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 11 |
Hb_001377_420 |
0.2010803088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001481_130 |
0.2018805558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011861_100 |
0.2040286066 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_000107_510 |
0.2061386182 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 15 |
Hb_002007_340 |
0.20702986 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_012163_020 |
0.2083387794 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000477_040 |
0.2142569777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002320_080 |
0.2144986025 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 19 |
Hb_025466_020 |
0.2159659258 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 20 |
Hb_075247_010 |
0.2189761668 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |