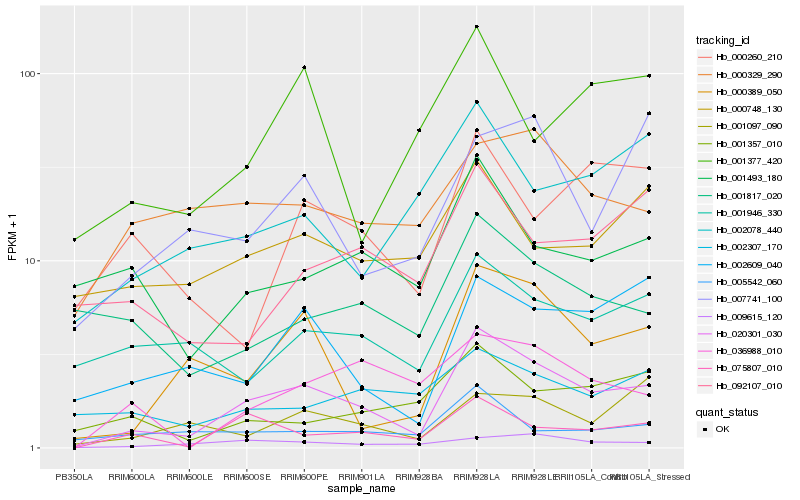
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020301_030 |
0.0 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 2 |
Hb_000389_050 |
0.2023407589 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 3 |
Hb_075807_010 |
0.2119593952 |
- |
- |
PREDICTED: uncharacterized protein LOC105631775 [Jatropha curcas] |
| 4 |
Hb_009615_120 |
0.2245603564 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 5 |
Hb_002609_040 |
0.2253901343 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 6 |
Hb_001946_330 |
0.2411111572 |
- |
- |
PREDICTED: transmembrane protein 128 [Jatropha curcas] |
| 7 |
Hb_036988_010 |
0.2430606283 |
- |
- |
hypothetical protein JCGZ_25496 [Jatropha curcas] |
| 8 |
Hb_002078_440 |
0.2446681879 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001493_180 |
0.244785925 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 10 |
Hb_005542_060 |
0.2454155599 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001357_010 |
0.2469486302 |
- |
- |
- |
| 12 |
Hb_000329_290 |
0.2491225143 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 13 |
Hb_007741_100 |
0.2492605218 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 14 |
Hb_001817_020 |
0.2509277256 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 15 |
Hb_000748_130 |
0.2513376598 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001097_090 |
0.251775048 |
- |
- |
Translation initiation factor SUI1 family protein, putative [Theobroma cacao] |
| 17 |
Hb_092107_010 |
0.2528790355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001377_420 |
0.2533252277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002307_170 |
0.2547213558 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 20 |
Hb_000260_210 |
0.2555491498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |