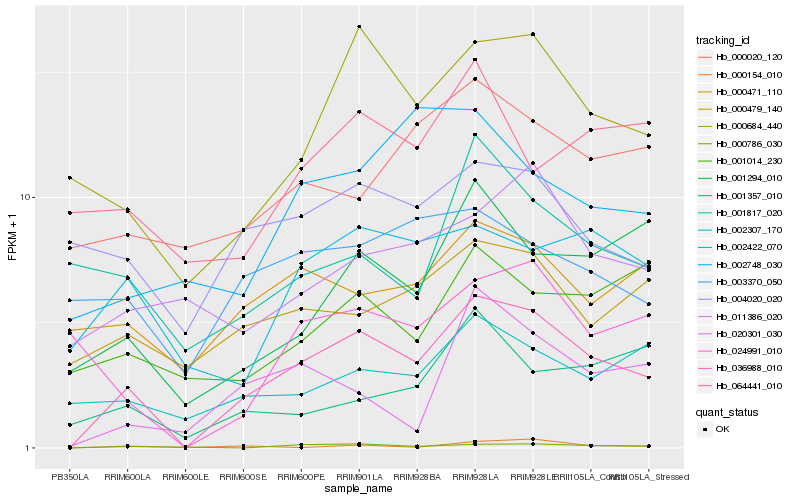
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036988_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_25496 [Jatropha curcas] |
| 2 |
Hb_000684_440 |
0.2080457455 |
- |
- |
hypothetical protein CICLE_v100111082mg, partial [Citrus clementina] |
| 3 |
Hb_001294_010 |
0.223254315 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 4 |
Hb_000786_030 |
0.2277268774 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 5 |
Hb_002422_070 |
0.2313067108 |
- |
- |
PREDICTED: putative protein TPRXL [Malus domestica] |
| 6 |
Hb_000479_140 |
0.2359553974 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 7 |
Hb_002307_170 |
0.2400991447 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 8 |
Hb_000471_110 |
0.2413794415 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |
| 9 |
Hb_020301_030 |
0.2430606283 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 10 |
Hb_004020_020 |
0.2449430523 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g65820 [Jatropha curcas] |
| 11 |
Hb_011386_020 |
0.2449847094 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000020_120 |
0.2456176254 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_002748_030 |
0.2462641458 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 14 |
Hb_024991_010 |
0.2464552243 |
- |
- |
hypothetical protein B456_007G217100 [Gossypium raimondii] |
| 15 |
Hb_003370_050 |
0.2485709154 |
- |
- |
PREDICTED: uncharacterized protein LOC103697550 [Phoenix dactylifera] |
| 16 |
Hb_001014_230 |
0.250322239 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 17 |
Hb_000154_010 |
0.2532512706 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_064441_010 |
0.2546083323 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001817_020 |
0.2554676691 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 20 |
Hb_001357_010 |
0.255667978 |
- |
- |
- |