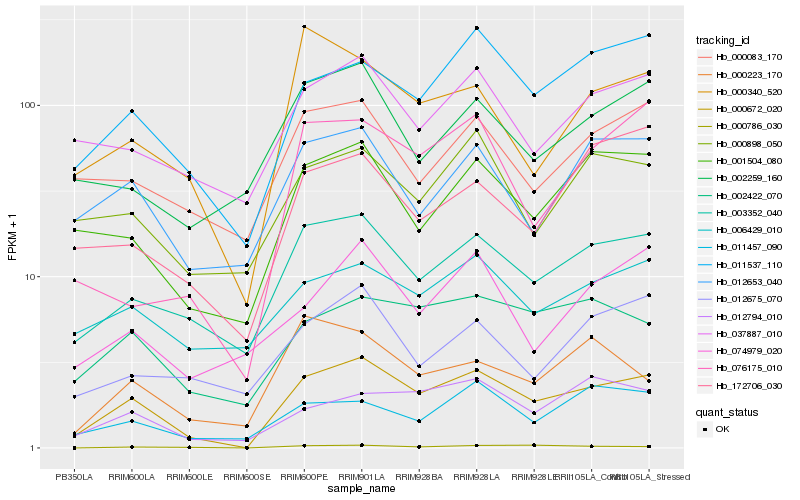
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000672_020 |
0.0 |
- |
- |
Peptidoglycan-binding LysM domain-containing protein, putative isoform 3 [Theobroma cacao] |
| 2 |
Hb_003352_040 |
0.1443721444 |
- |
- |
PREDICTED: 26S proteasome regulatory subunit RPN13 [Jatropha curcas] |
| 3 |
Hb_011537_110 |
0.1539516798 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 4 |
Hb_001504_080 |
0.1616928908 |
- |
- |
golgi snare 11 protein, putative [Ricinus communis] |
| 5 |
Hb_002422_070 |
0.1649029656 |
- |
- |
PREDICTED: putative protein TPRXL [Malus domestica] |
| 6 |
Hb_012794_010 |
0.1696944165 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 7 |
Hb_012653_040 |
0.1770773244 |
- |
- |
UV-induced protein uvi31, putative [Ricinus communis] |
| 8 |
Hb_011457_090 |
0.1779617425 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 9 |
Hb_037887_010 |
0.1794631369 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 10 |
Hb_000340_520 |
0.1794852326 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 11 |
Hb_006429_010 |
0.1839712367 |
- |
- |
PREDICTED: uncharacterized protein LOC105639724 [Jatropha curcas] |
| 12 |
Hb_000898_050 |
0.1851139657 |
- |
- |
- |
| 13 |
Hb_076175_010 |
0.1854783923 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB7 [Cucumis melo] |
| 14 |
Hb_000083_170 |
0.1879283735 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 7-like [Populus euphratica] |
| 15 |
Hb_002259_160 |
0.1889475811 |
- |
- |
hypothetical protein PSEUBRA_SCAF14g01508 [Pseudozyma brasiliensis GHG001] |
| 16 |
Hb_074979_020 |
0.1893136642 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000786_030 |
0.1894395555 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 18 |
Hb_000223_170 |
0.1905123579 |
- |
- |
- |
| 19 |
Hb_012675_070 |
0.1913342281 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Pyrus x bretschneideri] |
| 20 |
Hb_172706_030 |
0.1917163428 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |