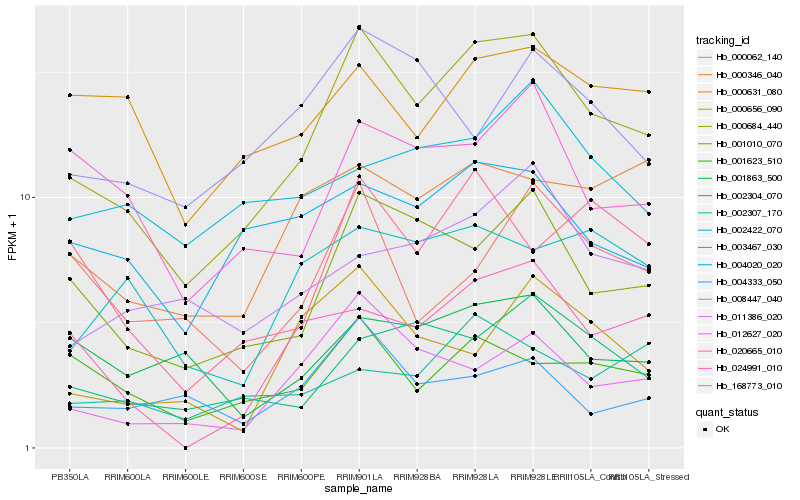
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_440 |
0.0 |
- |
- |
hypothetical protein CICLE_v100111082mg, partial [Citrus clementina] |
| 2 |
Hb_001010_070 |
0.1268236606 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 3 |
Hb_002304_070 |
0.1574367384 |
- |
- |
- |
| 4 |
Hb_012627_020 |
0.1580674451 |
- |
- |
PHYTOCHROME AND FLOWERING TIME 1 family protein [Populus trichocarpa] |
| 5 |
Hb_000346_040 |
0.1651917621 |
- |
- |
- |
| 6 |
Hb_020665_010 |
0.1656519351 |
- |
- |
PREDICTED: uncharacterized protein LOC103438977 isoform X2 [Malus domestica] |
| 7 |
Hb_004020_020 |
0.1667388823 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g65820 [Jatropha curcas] |
| 8 |
Hb_024991_010 |
0.1700024078 |
- |
- |
hypothetical protein B456_007G217100 [Gossypium raimondii] |
| 9 |
Hb_001623_510 |
0.1712961097 |
- |
- |
transporter, putative [Ricinus communis] |
| 10 |
Hb_168773_010 |
0.1733734875 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_011386_020 |
0.1775990866 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_002422_070 |
0.1787888854 |
- |
- |
PREDICTED: putative protein TPRXL [Malus domestica] |
| 13 |
Hb_008447_040 |
0.1821730945 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 14 |
Hb_001863_500 |
0.1838426158 |
- |
- |
Protein brittle-1, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_002307_170 |
0.1860763283 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 16 |
Hb_000631_080 |
0.1861505564 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Jatropha curcas] |
| 17 |
Hb_000656_090 |
0.1864310952 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 18 |
Hb_003467_030 |
0.1871440024 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 19 |
Hb_004333_050 |
0.187157126 |
- |
- |
- |
| 20 |
Hb_000062_140 |
0.1872413286 |
transcription factor |
TF Family: GRAS |
hypothetical protein JCGZ_17897 [Jatropha curcas] |