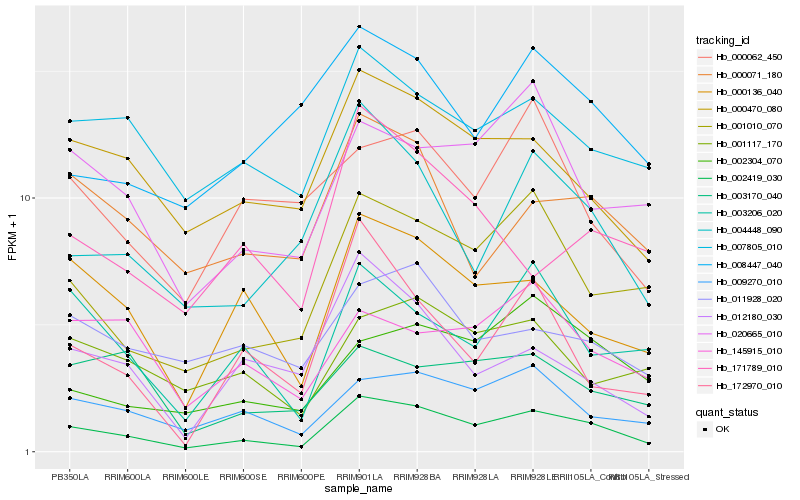
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002419_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_001010_070 |
0.1471233117 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 3 |
Hb_009270_010 |
0.1555038773 |
- |
- |
PREDICTED: uncharacterized protein LOC103421213 [Malus domestica] |
| 4 |
Hb_000136_040 |
0.1563468184 |
- |
- |
PREDICTED: protein RRP5 homolog [Jatropha curcas] |
| 5 |
Hb_004448_090 |
0.1627891771 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 6 |
Hb_000470_080 |
0.1677336077 |
- |
- |
Caldesmon, putative [Ricinus communis] |
| 7 |
Hb_003206_020 |
0.1705119136 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 8 |
Hb_020665_010 |
0.1762718065 |
- |
- |
PREDICTED: uncharacterized protein LOC103438977 isoform X2 [Malus domestica] |
| 9 |
Hb_002304_070 |
0.1810941542 |
- |
- |
- |
| 10 |
Hb_172970_010 |
0.1824859607 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 11 |
Hb_007805_010 |
0.1829627741 |
- |
- |
PREDICTED: ribosome biogenesis protein BMS1 homolog [Jatropha curcas] |
| 12 |
Hb_001117_170 |
0.1834122148 |
- |
- |
- |
| 13 |
Hb_012180_030 |
0.1884070945 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000071_180 |
0.1886670474 |
- |
- |
PREDICTED: putative DEAD-box ATP-dependent RNA helicase 29 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011928_020 |
0.1959938737 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |
| 16 |
Hb_145915_010 |
0.196030112 |
- |
- |
PREDICTED: uncharacterized protein LOC105630963 [Jatropha curcas] |
| 17 |
Hb_008447_040 |
0.1972843836 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 18 |
Hb_003170_040 |
0.1980562406 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 19 |
Hb_171789_010 |
0.1982524101 |
- |
- |
phosphoenolpyruvate carboxylase [Glycine max] |
| 20 |
Hb_000062_450 |
0.1992453542 |
- |
- |
hypothetical protein MTR_081s0009, partial [Medicago truncatula] |