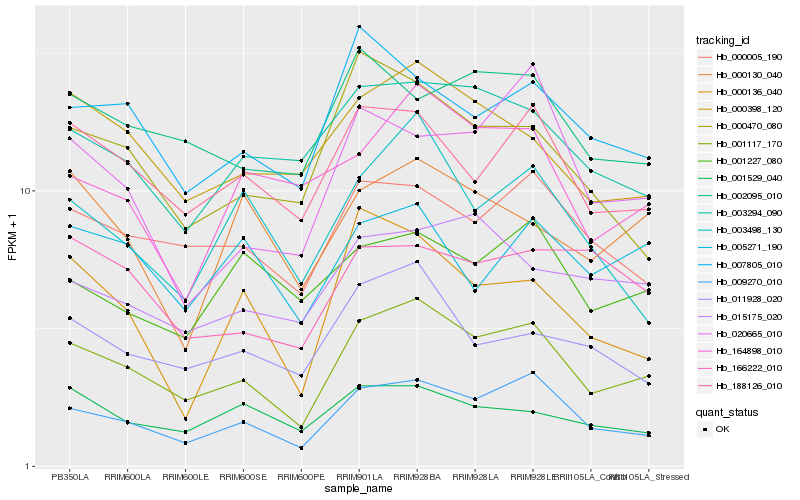
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_170 |
0.0 |
- |
- |
- |
| 2 |
Hb_009270_010 |
0.0741647982 |
- |
- |
PREDICTED: uncharacterized protein LOC103421213 [Malus domestica] |
| 3 |
Hb_188126_010 |
0.1082274868 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like isoform X3 [Jatropha curcas] |
| 4 |
Hb_000005_190 |
0.1134345799 |
- |
- |
hypothetical protein JCGZ_05438 [Jatropha curcas] |
| 5 |
Hb_000398_120 |
0.1175585288 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3-like isoform X4 [Gossypium raimondii] |
| 6 |
Hb_007805_010 |
0.1201274473 |
- |
- |
PREDICTED: ribosome biogenesis protein BMS1 homolog [Jatropha curcas] |
| 7 |
Hb_011928_020 |
0.122894727 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |
| 8 |
Hb_003294_090 |
0.1231825056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000470_080 |
0.1280859973 |
- |
- |
Caldesmon, putative [Ricinus communis] |
| 10 |
Hb_002095_010 |
0.1282210095 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003498_130 |
0.1298533411 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |
| 12 |
Hb_001529_040 |
0.1317901305 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 13 |
Hb_015175_020 |
0.1327193794 |
- |
- |
hypothetical protein JCGZ_14452 [Jatropha curcas] |
| 14 |
Hb_020665_010 |
0.1347954437 |
- |
- |
PREDICTED: uncharacterized protein LOC103438977 isoform X2 [Malus domestica] |
| 15 |
Hb_164898_010 |
0.1362065829 |
- |
- |
Cysteine-rich protein, putative [Theobroma cacao] |
| 16 |
Hb_005271_190 |
0.1366434942 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 17 |
Hb_000136_040 |
0.1386726102 |
- |
- |
PREDICTED: protein RRP5 homolog [Jatropha curcas] |
| 18 |
Hb_001227_080 |
0.1387180551 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_166222_010 |
0.1403525888 |
- |
- |
hypothetical protein POPTR_0017s08350g [Populus trichocarpa] |
| 20 |
Hb_000130_040 |
0.1407143183 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |