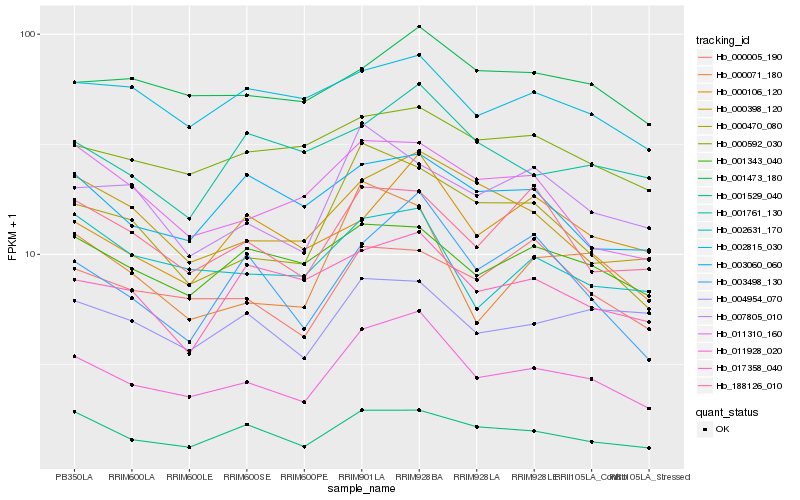
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011928_020 |
0.0 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |
| 2 |
Hb_001529_040 |
0.092870186 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 3 |
Hb_000470_080 |
0.1002821185 |
- |
- |
Caldesmon, putative [Ricinus communis] |
| 4 |
Hb_000398_120 |
0.1045468305 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3-like isoform X4 [Gossypium raimondii] |
| 5 |
Hb_003498_130 |
0.108888074 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |
| 6 |
Hb_002631_170 |
0.1090811463 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 7 |
Hb_000071_180 |
0.1096562671 |
- |
- |
PREDICTED: putative DEAD-box ATP-dependent RNA helicase 29 isoform X1 [Jatropha curcas] |
| 8 |
Hb_188126_010 |
0.1111161285 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_001473_180 |
0.1119440312 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 10 |
Hb_001343_040 |
0.1127767296 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 11 |
Hb_000106_120 |
0.1130567256 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 12 |
Hb_011310_160 |
0.1133264936 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 13 |
Hb_000592_030 |
0.1136640078 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 14 |
Hb_000005_190 |
0.114397785 |
- |
- |
hypothetical protein JCGZ_05438 [Jatropha curcas] |
| 15 |
Hb_017358_040 |
0.1145182284 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 16 |
Hb_001761_130 |
0.1151382806 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007805_010 |
0.1158164735 |
- |
- |
PREDICTED: ribosome biogenesis protein BMS1 homolog [Jatropha curcas] |
| 18 |
Hb_004954_070 |
0.1163843406 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 19 |
Hb_003060_060 |
0.1166102318 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 20 |
Hb_002815_030 |
0.1171155862 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |