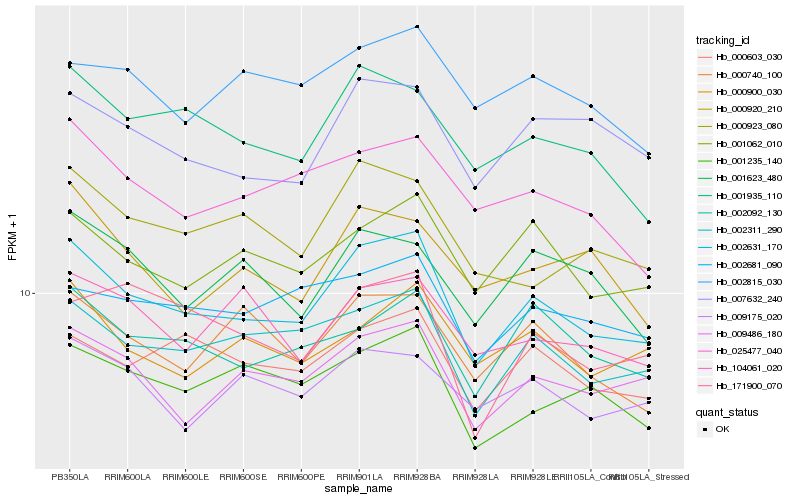
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_170 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 2 |
Hb_002311_290 |
0.0645424246 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 3 |
Hb_009486_180 |
0.0726925692 |
- |
- |
unknown [Glycine max] |
| 4 |
Hb_001062_010 |
0.0729128808 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 5 |
Hb_001935_110 |
0.0743123935 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 6 |
Hb_000603_030 |
0.0762010905 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 7 |
Hb_002092_130 |
0.0769675507 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 8 |
Hb_007632_240 |
0.0796156533 |
- |
- |
transporter, putative [Ricinus communis] |
| 9 |
Hb_001235_140 |
0.0802677871 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 10 |
Hb_025477_040 |
0.0804441374 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 11 |
Hb_000923_080 |
0.0823953361 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 12 |
Hb_002681_090 |
0.0832195505 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000740_100 |
0.0832647594 |
- |
- |
calpain, putative [Ricinus communis] |
| 14 |
Hb_000900_030 |
0.0833842883 |
- |
- |
PREDICTED: WD repeat-containing protein 3 [Jatropha curcas] |
| 15 |
Hb_009175_020 |
0.0862898998 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_001623_480 |
0.0896540944 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 17 |
Hb_000920_210 |
0.0898478159 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 18 |
Hb_104061_020 |
0.0901928636 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 19 |
Hb_002815_030 |
0.0907414163 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 20 |
Hb_171900_070 |
0.0911819271 |
- |
- |
- |