| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
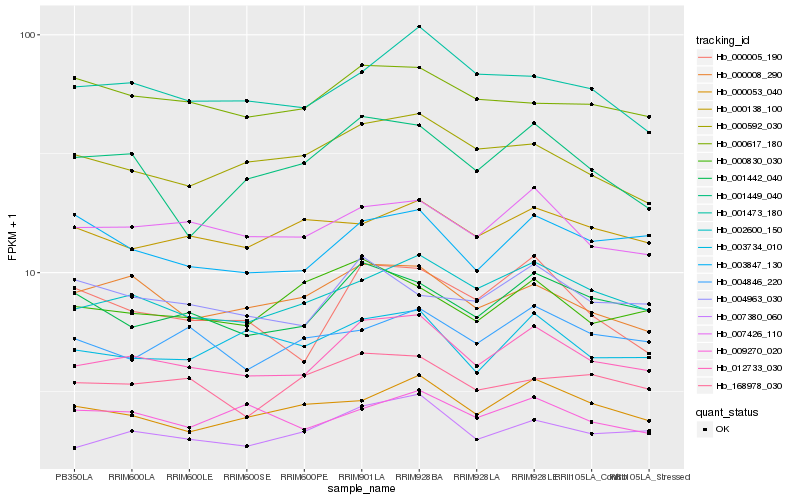
Hb_012733_030 |
0.0 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |
| 2 |
Hb_007380_060 |
0.0516035848 |
- |
- |
hypothetical protein POPTR_0006s00280g [Populus trichocarpa] |
| 3 |
Hb_002600_150 |
0.0533061931 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007426_110 |
0.0624995389 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_001442_040 |
0.063839815 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000008_290 |
0.0700586261 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 7 |
Hb_000053_040 |
0.0721105389 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 8 |
Hb_003734_010 |
0.072476388 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 9 |
Hb_000592_030 |
0.0735071691 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 10 |
Hb_001473_180 |
0.0739269759 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 11 |
Hb_168978_030 |
0.0790743614 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 12 |
Hb_003847_130 |
0.0790766811 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 13 |
Hb_009270_020 |
0.080483341 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 14 |
Hb_004963_030 |
0.0808060021 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_000617_180 |
0.0808966428 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 16 |
Hb_001449_040 |
0.081374023 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000005_190 |
0.0814674098 |
- |
- |
hypothetical protein JCGZ_05438 [Jatropha curcas] |
| 18 |
Hb_004846_220 |
0.0816879466 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000138_100 |
0.0826075276 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 20 |
Hb_000830_030 |
0.0827873356 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |