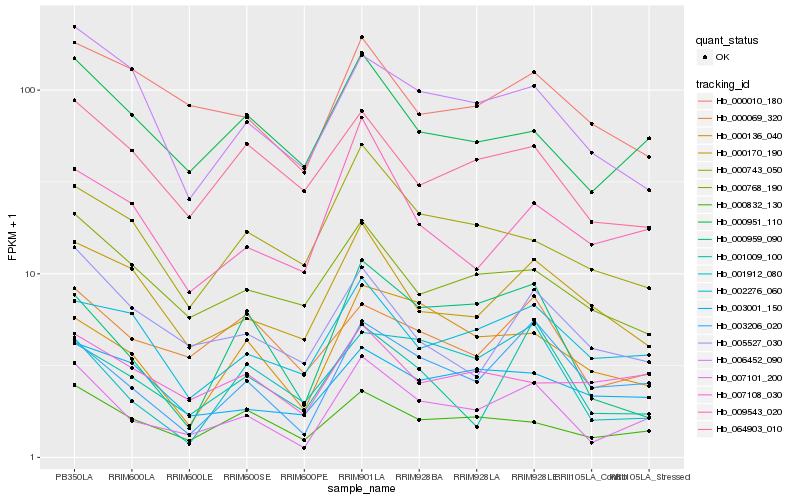
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_200 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003206_020 |
0.1420174827 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 3 |
Hb_000832_130 |
0.1596343448 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g39620 [Jatropha curcas] |
| 4 |
Hb_000951_110 |
0.1662600922 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 5 |
Hb_000959_090 |
0.177084556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000136_040 |
0.1771216566 |
- |
- |
PREDICTED: protein RRP5 homolog [Jatropha curcas] |
| 7 |
Hb_009543_020 |
0.1772322501 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 8 |
Hb_002276_060 |
0.1772348608 |
- |
- |
hypothetical protein CISIN_1g002019mg [Citrus sinensis] |
| 9 |
Hb_000069_320 |
0.179782825 |
- |
- |
PREDICTED: uncharacterized protein LOC105642846 [Jatropha curcas] |
| 10 |
Hb_001009_100 |
0.182782396 |
- |
- |
- |
| 11 |
Hb_000170_190 |
0.1828662307 |
- |
- |
PREDICTED: uncharacterized protein LOC105638988 [Jatropha curcas] |
| 12 |
Hb_006452_090 |
0.1843434279 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 5B [Jatropha curcas] |
| 13 |
Hb_005527_030 |
0.1860730527 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g42630 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000743_050 |
0.1860958372 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_001912_080 |
0.1874909899 |
- |
- |
vacuolar protein sorting-associated protein, putative [Ricinus communis] |
| 16 |
Hb_000768_190 |
0.1875197857 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 17 |
Hb_064903_010 |
0.1926050512 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 18 |
Hb_007108_030 |
0.1926656422 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000010_180 |
0.1931639877 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_003001_150 |
0.1934120507 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |