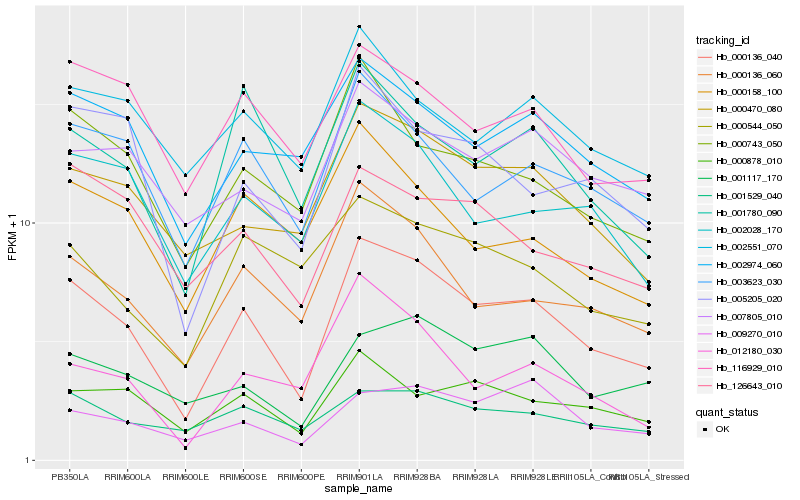
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000136_040 |
0.0 |
- |
- |
PREDICTED: protein RRP5 homolog [Jatropha curcas] |
| 2 |
Hb_000136_060 |
0.1177715909 |
- |
- |
hypothetical protein POPTR_0016s06250g [Populus trichocarpa] |
| 3 |
Hb_003623_030 |
0.1212778939 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 14 [Jatropha curcas] |
| 4 |
Hb_000470_080 |
0.1236836154 |
- |
- |
Caldesmon, putative [Ricinus communis] |
| 5 |
Hb_000743_050 |
0.1278850506 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_005205_020 |
0.1294225911 |
- |
- |
PREDICTED: protein FAM135B-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_002028_170 |
0.1297269921 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 8 |
Hb_001780_090 |
0.1319009813 |
- |
- |
PREDICTED: uncharacterized protein LOC105631099 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000878_010 |
0.1326766255 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g06400, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002974_060 |
0.1330227315 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 11 |
Hb_001529_040 |
0.1369479737 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 12 |
Hb_116929_010 |
0.1383909076 |
- |
- |
PREDICTED: nuclear-pore anchor isoform X2 [Jatropha curcas] |
| 13 |
Hb_001117_170 |
0.1386726102 |
- |
- |
- |
| 14 |
Hb_007805_010 |
0.1410667057 |
- |
- |
PREDICTED: ribosome biogenesis protein BMS1 homolog [Jatropha curcas] |
| 15 |
Hb_009270_010 |
0.1420549338 |
- |
- |
PREDICTED: uncharacterized protein LOC103421213 [Malus domestica] |
| 16 |
Hb_126643_010 |
0.1432668134 |
- |
- |
Protein transport protein Sec24C, putative [Ricinus communis] |
| 17 |
Hb_000158_100 |
0.1447275239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002551_070 |
0.1447815706 |
- |
- |
hypothetical protein POPTR_0019s10130g [Populus trichocarpa] |
| 19 |
Hb_000544_050 |
0.1455238719 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 20 |
Hb_012180_030 |
0.1455734665 |
- |
- |
unnamed protein product [Vitis vinifera] |