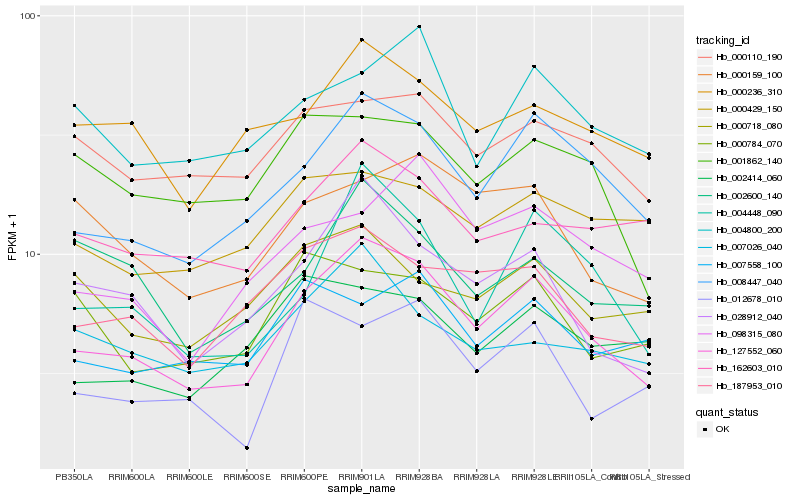
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127552_060 |
0.0 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_008447_040 |
0.0865786815 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 3 |
Hb_028912_040 |
0.1091202481 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |
| 4 |
Hb_004448_090 |
0.1127528387 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 5 |
Hb_187953_010 |
0.1209547441 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000236_310 |
0.1345216039 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Malus domestica] |
| 7 |
Hb_000159_100 |
0.1365191252 |
- |
- |
PREDICTED: uncharacterized protein At4g19900 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000110_190 |
0.1386473386 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 9 |
Hb_162603_010 |
0.1402296707 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_001862_140 |
0.1409156317 |
- |
- |
natural resistance-associated macrophage protein, putative [Ricinus communis] |
| 11 |
Hb_098315_080 |
0.1410382432 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_007026_040 |
0.1416815224 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |
| 13 |
Hb_004800_200 |
0.1424674188 |
- |
- |
- |
| 14 |
Hb_002600_140 |
0.1434192454 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 15 |
Hb_002414_060 |
0.1448606665 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 16 |
Hb_000784_070 |
0.1480720637 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_007558_100 |
0.1481886392 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 18 |
Hb_012678_010 |
0.1484091767 |
- |
- |
PREDICTED: outer envelope protein 61-like [Gossypium raimondii] |
| 19 |
Hb_000429_150 |
0.1485660754 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 20 |
Hb_000718_080 |
0.1491093601 |
- |
- |
hypothetical protein PRUPE_ppa005259mg [Prunus persica] |