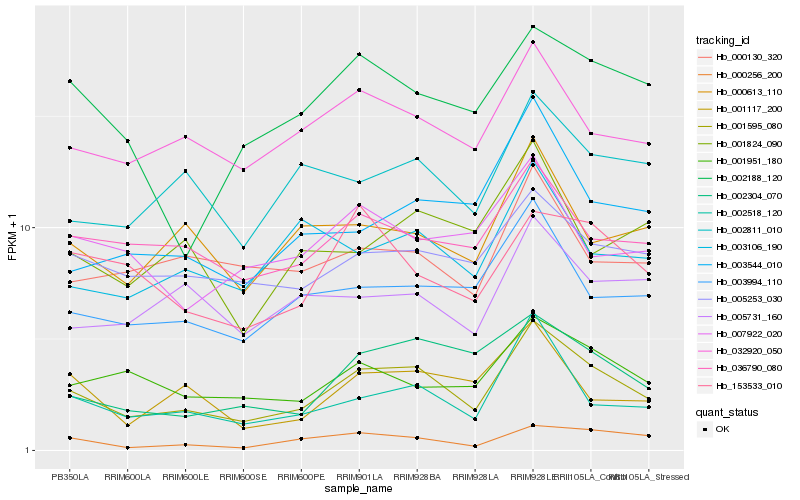
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001595_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002518_120 |
0.1312761525 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |
| 3 |
Hb_032920_050 |
0.1367876639 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003994_110 |
0.1383392182 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 5 |
Hb_002304_070 |
0.1396450314 |
- |
- |
- |
| 6 |
Hb_000256_200 |
0.141891631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001951_180 |
0.1441460079 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_003544_010 |
0.1487891231 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_036790_080 |
0.1488919379 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 10 |
Hb_005253_030 |
0.1497814053 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002811_010 |
0.1508662323 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 12 |
Hb_000613_110 |
0.1522271688 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 13 |
Hb_001117_200 |
0.1526394389 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002188_120 |
0.1559884157 |
- |
- |
PREDICTED: thymocyte nuclear protein 1 [Pyrus x bretschneideri] |
| 15 |
Hb_000130_320 |
0.1583198639 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 16 |
Hb_153533_010 |
0.1586821178 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X3 [Jatropha curcas] |
| 17 |
Hb_003106_190 |
0.1598219125 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 18 |
Hb_001824_090 |
0.1607954373 |
- |
- |
- |
| 19 |
Hb_005731_160 |
0.1608056045 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_007922_020 |
0.1624544649 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |