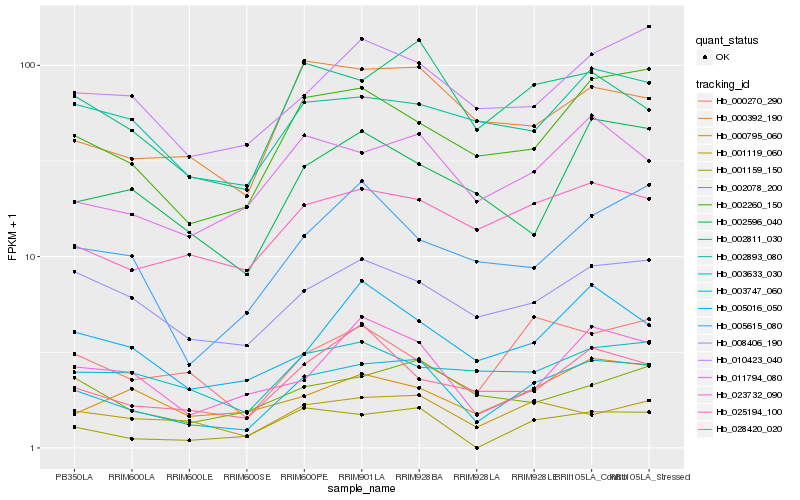
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003747_060 |
0.0 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 2 isoform X2 [Cucumis sativus] |
| 2 |
Hb_002260_150 |
0.1230136925 |
- |
- |
PREDICTED: uncharacterized protein LOC105647187 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002811_030 |
0.1294874253 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 4 |
Hb_011794_080 |
0.135015152 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 5 |
Hb_001119_060 |
0.1372077016 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X3 [Malus domestica] |
| 6 |
Hb_008406_190 |
0.14198113 |
- |
- |
PREDICTED: RNA polymerase II subunit 5-mediating protein homolog isoform X2 [Jatropha curcas] |
| 7 |
Hb_010423_040 |
0.142199547 |
- |
- |
hypothetical protein JCGZ_17463 [Jatropha curcas] |
| 8 |
Hb_025194_100 |
0.1427000555 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 9 |
Hb_023732_090 |
0.1435907991 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_000392_190 |
0.1447469663 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 11 |
Hb_005016_050 |
0.1478353813 |
- |
- |
- |
| 12 |
Hb_001159_150 |
0.1495489283 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_002078_200 |
0.1508113363 |
- |
- |
Mitochondrial import receptor subunit TOM20, putative [Ricinus communis] |
| 14 |
Hb_002893_080 |
0.1543821152 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |
| 15 |
Hb_000270_290 |
0.1553163525 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 16 |
Hb_028420_020 |
0.1562182227 |
- |
- |
PREDICTED: uncharacterized protein LOC105647566 [Jatropha curcas] |
| 17 |
Hb_002596_040 |
0.1571601995 |
- |
- |
PREDICTED: uncharacterized protein LOC105645264 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003633_030 |
0.1576220192 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 19 |
Hb_000795_060 |
0.1580558193 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 20 |
Hb_005615_080 |
0.1589450838 |
- |
- |
PREDICTED: uncharacterized protein LOC105649609 [Jatropha curcas] |