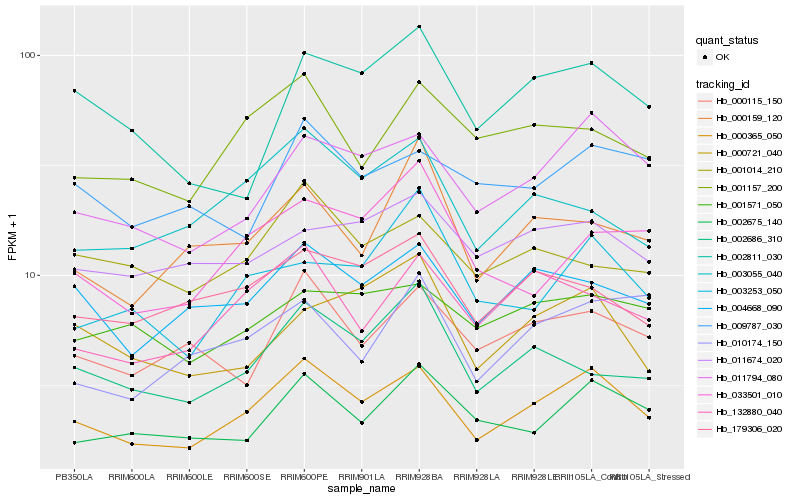
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_011794_080 |
0.0893444932 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 3 |
Hb_004668_090 |
0.1166620933 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 4 |
Hb_132880_040 |
0.1178991658 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_002675_140 |
0.1195797322 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 6 |
Hb_000115_150 |
0.1237590439 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 7 |
Hb_001157_200 |
0.1256739604 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 8 |
Hb_002811_030 |
0.1319127279 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 9 |
Hb_003055_040 |
0.1331571332 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_179306_020 |
0.1332819929 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009787_030 |
0.1361354931 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000721_040 |
0.1364453736 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002686_310 |
0.1365394621 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 14 |
Hb_001571_050 |
0.1368001991 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3-like [Populus euphratica] |
| 15 |
Hb_033501_010 |
0.1374494837 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 16 |
Hb_001014_210 |
0.1393841024 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 17 |
Hb_010174_150 |
0.1402151181 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_000159_120 |
0.1403777033 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 19 |
Hb_011674_020 |
0.1407652206 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 20 |
Hb_003253_050 |
0.1409186562 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |