| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
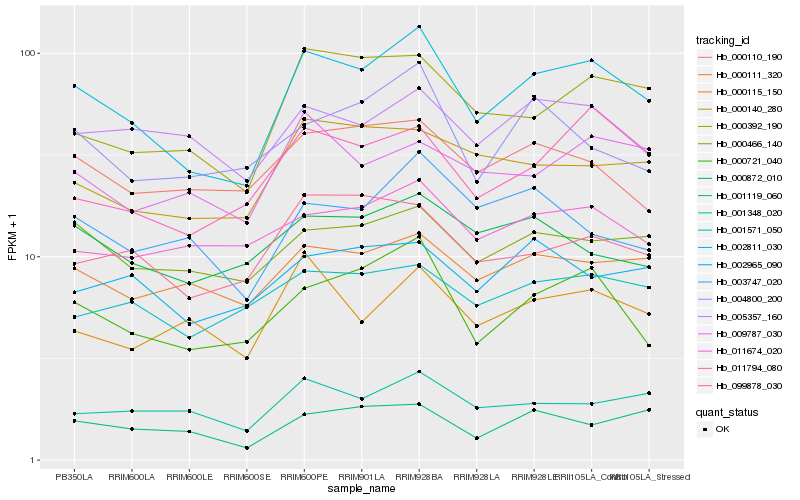
Hb_002811_030 |
0.0 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 2 |
Hb_000721_040 |
0.0953710213 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000392_190 |
0.1067253146 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 4 |
Hb_011794_080 |
0.1127711764 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 5 |
Hb_000466_140 |
0.1144391535 |
- |
- |
hypothetical protein JCGZ_02122 [Jatropha curcas] |
| 6 |
Hb_001348_020 |
0.1162637184 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000111_320 |
0.1169940835 |
transcription factor |
TF Family: bZIP |
Transcription factor HBP-1b(c1), putative [Ricinus communis] |
| 8 |
Hb_099878_030 |
0.1182196851 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 9 |
Hb_000140_280 |
0.1183151912 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 10 |
Hb_005357_160 |
0.123099221 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 11 |
Hb_000115_150 |
0.1242604004 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 12 |
Hb_001119_060 |
0.124896086 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X3 [Malus domestica] |
| 13 |
Hb_000110_190 |
0.1249049509 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 14 |
Hb_001571_050 |
0.1253690153 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3-like [Populus euphratica] |
| 15 |
Hb_004800_200 |
0.1263035232 |
- |
- |
- |
| 16 |
Hb_011674_020 |
0.126329045 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 17 |
Hb_000872_010 |
0.126572799 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 18 |
Hb_003747_020 |
0.1272767224 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X1 [Jatropha curcas] |
| 19 |
Hb_002965_090 |
0.1274411041 |
- |
- |
importin alpha, putative [Ricinus communis] |
| 20 |
Hb_009787_030 |
0.1274600718 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |