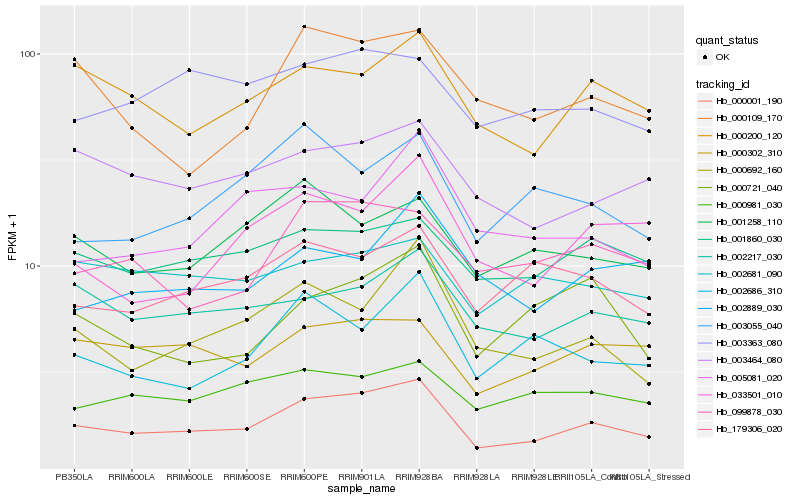
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000001_190 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0082522mg, partial [Citrus sinensis] |
| 2 |
Hb_000200_120 |
0.1100702352 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002217_030 |
0.1110293981 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000692_160 |
0.1117776606 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 5 |
Hb_003464_080 |
0.1154761708 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 6 |
Hb_000302_310 |
0.1162470253 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 7 |
Hb_099878_030 |
0.1170324729 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 8 |
Hb_001860_030 |
0.1174038241 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 9 |
Hb_179306_020 |
0.117706243 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002686_310 |
0.1206886155 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 11 |
Hb_000981_030 |
0.1224026838 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 12 |
Hb_001258_110 |
0.122409369 |
- |
- |
transporter, putative [Ricinus communis] |
| 13 |
Hb_033501_010 |
0.1237954335 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 14 |
Hb_000109_170 |
0.1238472041 |
- |
- |
PREDICTED: uncharacterized protein LOC105649868 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003363_080 |
0.1257107024 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 16 |
Hb_000721_040 |
0.1282097171 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002681_090 |
0.1295027561 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003055_040 |
0.1296686156 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005081_020 |
0.130072836 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 20 |
Hb_002889_030 |
0.1304913723 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |