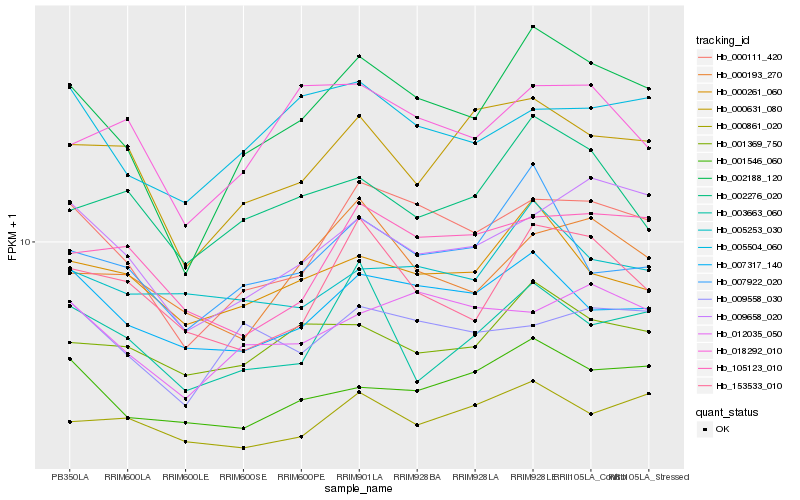
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002188_120 |
0.0 |
- |
- |
PREDICTED: thymocyte nuclear protein 1 [Pyrus x bretschneideri] |
| 2 |
Hb_000111_420 |
0.0905089267 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Populus euphratica] |
| 3 |
Hb_007922_020 |
0.120823351 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 4 |
Hb_001546_060 |
0.1242109747 |
- |
- |
PREDICTED: uncharacterized protein LOC105133943 isoform X4 [Populus euphratica] |
| 5 |
Hb_000631_080 |
0.1244076721 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Jatropha curcas] |
| 6 |
Hb_001369_750 |
0.1251233329 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 7 |
Hb_009658_020 |
0.1262557873 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 8 |
Hb_000261_060 |
0.1264936831 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 9 |
Hb_153533_010 |
0.1267802589 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X3 [Jatropha curcas] |
| 10 |
Hb_007317_140 |
0.1278522986 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 11 |
Hb_002276_020 |
0.1325533444 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 12 |
Hb_012035_050 |
0.1355456581 |
- |
- |
hypothetical protein JCGZ_10788 [Jatropha curcas] |
| 13 |
Hb_000193_270 |
0.1368541922 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_018292_010 |
0.1376134318 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Jatropha curcas] |
| 15 |
Hb_000861_020 |
0.1381253968 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 16 |
Hb_005504_060 |
0.1381375148 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Populus euphratica] |
| 17 |
Hb_003663_060 |
0.139415533 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g43820 [Jatropha curcas] |
| 18 |
Hb_105123_010 |
0.1398017703 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 19 |
Hb_009558_030 |
0.140371141 |
- |
- |
myb3r3, putative [Ricinus communis] |
| 20 |
Hb_005253_030 |
0.1430262718 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |