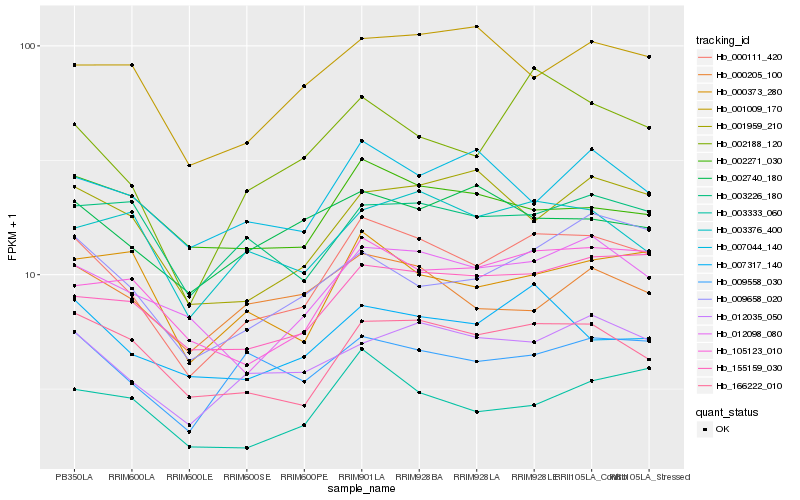
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_420 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Populus euphratica] |
| 2 |
Hb_012035_050 |
0.0891860915 |
- |
- |
hypothetical protein JCGZ_10788 [Jatropha curcas] |
| 3 |
Hb_002188_120 |
0.0905089267 |
- |
- |
PREDICTED: thymocyte nuclear protein 1 [Pyrus x bretschneideri] |
| 4 |
Hb_166222_010 |
0.0955360084 |
- |
- |
hypothetical protein POPTR_0017s08350g [Populus trichocarpa] |
| 5 |
Hb_105123_010 |
0.0966565062 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 6 |
Hb_155159_030 |
0.0971665416 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 7 |
Hb_012098_080 |
0.1005236881 |
- |
- |
KOM, putative [Ricinus communis] |
| 8 |
Hb_007317_140 |
0.1023745664 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 9 |
Hb_009558_030 |
0.1033415239 |
- |
- |
myb3r3, putative [Ricinus communis] |
| 10 |
Hb_003333_060 |
0.1078309958 |
- |
- |
PREDICTED: uncharacterized protein LOC105628461 [Jatropha curcas] |
| 11 |
Hb_009658_020 |
0.1080218094 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 12 |
Hb_000373_280 |
0.1103815551 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001959_210 |
0.111950313 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 14 |
Hb_001009_170 |
0.1153362731 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 15 |
Hb_003376_400 |
0.1153380663 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 16 |
Hb_003226_180 |
0.1153619279 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 17 |
Hb_007044_140 |
0.1188168164 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta [Jatropha curcas] |
| 18 |
Hb_002740_180 |
0.1200451129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000205_100 |
0.1204815686 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 20 |
Hb_002271_030 |
0.1206980569 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |