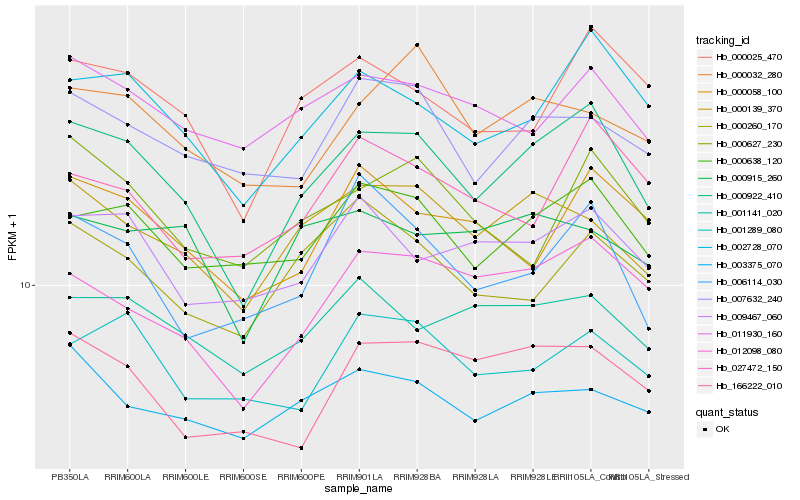
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_410 |
0.0 |
- |
- |
PREDICTED: anthranilate phosphoribosyltransferase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002728_070 |
0.0775726879 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 3 |
Hb_012098_080 |
0.0906803338 |
- |
- |
KOM, putative [Ricinus communis] |
| 4 |
Hb_000025_470 |
0.0913111465 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 5 |
Hb_000638_120 |
0.0918061491 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 2, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_000139_370 |
0.0927820296 |
- |
- |
ubiquitin-activating enzyme E1c, putative [Ricinus communis] |
| 7 |
Hb_009467_060 |
0.1048852277 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007632_240 |
0.1064824935 |
- |
- |
transporter, putative [Ricinus communis] |
| 9 |
Hb_001141_020 |
0.1071603498 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 10 |
Hb_011930_160 |
0.107388341 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 38-like [Jatropha curcas] |
| 11 |
Hb_000058_100 |
0.108061736 |
- |
- |
PREDICTED: RNA-directed DNA methylation 4 [Jatropha curcas] |
| 12 |
Hb_003375_070 |
0.1091596348 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 13 |
Hb_001289_080 |
0.109332396 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 14 |
Hb_166222_010 |
0.1094840407 |
- |
- |
hypothetical protein POPTR_0017s08350g [Populus trichocarpa] |
| 15 |
Hb_006114_030 |
0.1106144349 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 16 |
Hb_000260_170 |
0.1107229187 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 17 |
Hb_000627_230 |
0.1124451769 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_027472_150 |
0.1138320933 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 10 [Jatropha curcas] |
| 19 |
Hb_000032_280 |
0.1142760556 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 20 |
Hb_000915_260 |
0.1145706961 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |