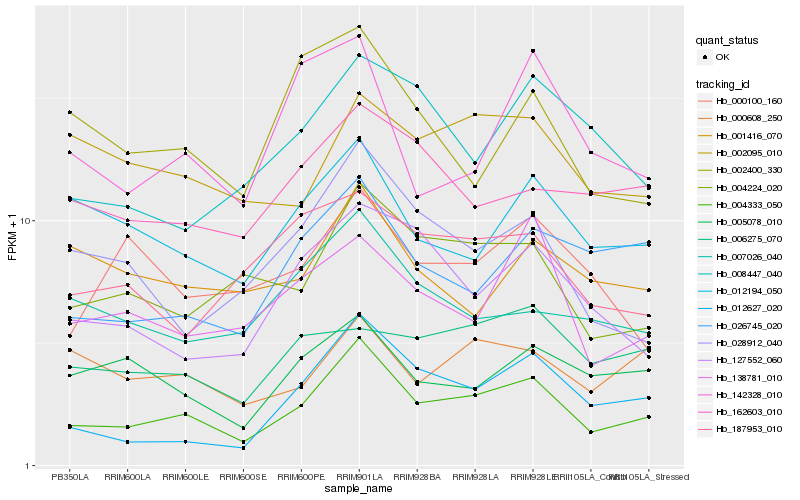
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004333_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_004224_020 |
0.141131681 |
- |
- |
PREDICTED: uncharacterized protein LOC105643786 [Jatropha curcas] |
| 3 |
Hb_012627_020 |
0.1482057982 |
- |
- |
PHYTOCHROME AND FLOWERING TIME 1 family protein [Populus trichocarpa] |
| 4 |
Hb_026745_020 |
0.1518877221 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 5 |
Hb_028912_040 |
0.1549264697 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |
| 6 |
Hb_127552_060 |
0.1561008135 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_012194_050 |
0.1563748517 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 8 |
Hb_138781_010 |
0.1578546833 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 9 |
Hb_005078_010 |
0.1620391421 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 10 |
Hb_162603_010 |
0.162741696 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_007026_040 |
0.1633251442 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |
| 12 |
Hb_008447_040 |
0.1678207223 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 13 |
Hb_000608_250 |
0.168043075 |
- |
- |
Cytidine/deoxycytidylate deaminase family protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_002095_010 |
0.1685581686 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_142328_010 |
0.1688219898 |
- |
- |
PREDICTED: uncharacterized protein LOC105630513 isoform X1 [Jatropha curcas] |
| 16 |
Hb_187953_010 |
0.1696408222 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006275_070 |
0.1699359559 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 18 |
Hb_002400_330 |
0.1728575794 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_000100_160 |
0.1738631156 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 20 |
Hb_001416_070 |
0.1747301455 |
- |
- |
oxidoreductase, putative [Ricinus communis] |