| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
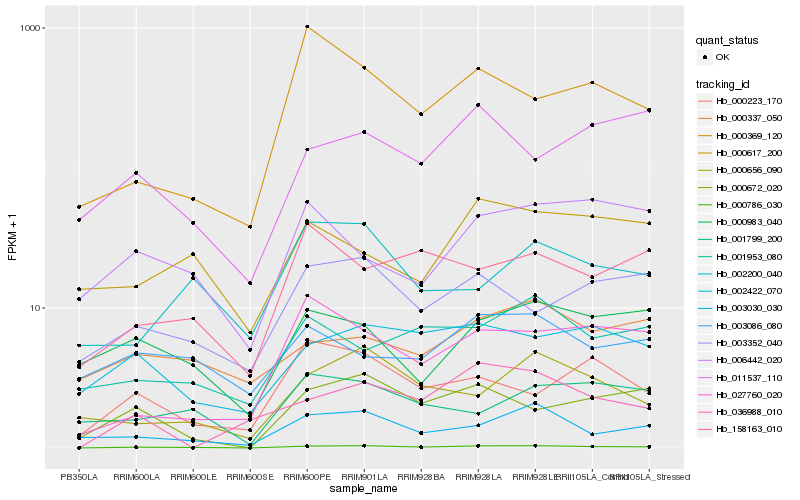
Hb_000786_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 2 |
Hb_000672_020 |
0.1894395555 |
- |
- |
Peptidoglycan-binding LysM domain-containing protein, putative isoform 3 [Theobroma cacao] |
| 3 |
Hb_003030_030 |
0.20447571 |
- |
- |
- |
| 4 |
Hb_002422_070 |
0.208960431 |
- |
- |
PREDICTED: putative protein TPRXL [Malus domestica] |
| 5 |
Hb_027760_020 |
0.2105890119 |
- |
- |
hypothetical protein M569_04462, partial [Genlisea aurea] |
| 6 |
Hb_000656_090 |
0.2151522655 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 7 |
Hb_003352_040 |
0.2175782981 |
- |
- |
PREDICTED: 26S proteasome regulatory subunit RPN13 [Jatropha curcas] |
| 8 |
Hb_000983_040 |
0.2249241963 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 9 |
Hb_000223_170 |
0.2257699229 |
- |
- |
- |
| 10 |
Hb_158163_010 |
0.2259813928 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_036988_010 |
0.2277268774 |
- |
- |
hypothetical protein JCGZ_25496 [Jatropha curcas] |
| 12 |
Hb_001799_200 |
0.2292781518 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 13 |
Hb_000369_120 |
0.2295674918 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |
| 14 |
Hb_001953_080 |
0.2300273522 |
- |
- |
pseudouridine synthase family protein, partial [Manihot esculenta] |
| 15 |
Hb_011537_110 |
0.2358418057 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 16 |
Hb_002200_040 |
0.236472145 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006442_020 |
0.2378404608 |
- |
- |
PREDICTED: very-long-chain enoyl-CoA reductase [Jatropha curcas] |
| 18 |
Hb_000617_200 |
0.2381032137 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 19 |
Hb_000337_050 |
0.239208056 |
- |
- |
PREDICTED: chloroplast envelope membrane protein [Jatropha curcas] |
| 20 |
Hb_003086_080 |
0.2426357629 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |