| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
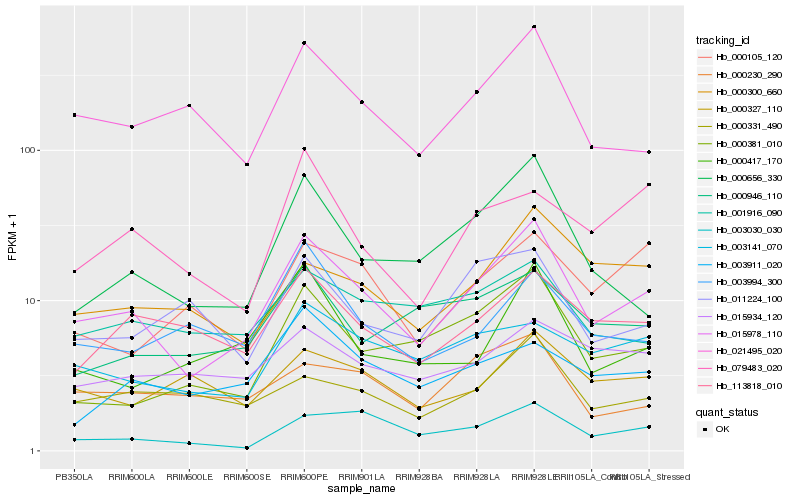
Hb_015978_110 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g62470, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_021495_020 |
0.1549249557 |
- |
- |
chitinase, putative [Ricinus communis] |
| 3 |
Hb_000656_330 |
0.1597706158 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 4 |
Hb_000381_010 |
0.1664392733 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_003030_030 |
0.1671830162 |
- |
- |
- |
| 6 |
Hb_113818_010 |
0.1676724995 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 7 |
Hb_000417_170 |
0.1720224891 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 8 |
Hb_003141_070 |
0.1725940833 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 9 |
Hb_000331_490 |
0.1726565625 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000300_660 |
0.1740709098 |
- |
- |
unknown [Glycine max] |
| 11 |
Hb_001916_090 |
0.1782693383 |
- |
- |
beta-1,2-n-acetylglucosaminyltransferase II, putative [Ricinus communis] |
| 12 |
Hb_000230_290 |
0.1784863241 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_015934_120 |
0.1833235199 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 14 |
Hb_000946_110 |
0.1852185842 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 15 |
Hb_003994_300 |
0.185415409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000327_110 |
0.1871420481 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 17 |
Hb_003911_020 |
0.1888223503 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 18 |
Hb_000105_120 |
0.1907417835 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 19 |
Hb_011224_100 |
0.191370014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_079483_020 |
0.1920463383 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X2 [Jatropha curcas] |