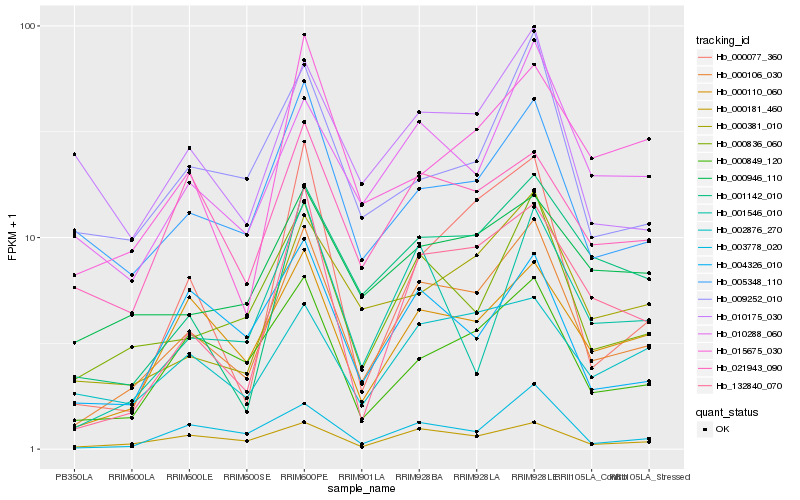
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000106_030 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_000836_060 |
0.11976645 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 3 |
Hb_000181_460 |
0.1254847365 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 4 |
Hb_132840_070 |
0.1316966275 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 5 |
Hb_000110_060 |
0.1363356388 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 6 |
Hb_000849_120 |
0.1392444646 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 7 |
Hb_003778_020 |
0.142788293 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 8 |
Hb_000381_010 |
0.1432041345 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_005348_110 |
0.1490581649 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 10 |
Hb_001142_010 |
0.1538078541 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |
| 11 |
Hb_010288_060 |
0.154823261 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 12 |
Hb_000077_360 |
0.1618137315 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 13 |
Hb_009252_010 |
0.1629987542 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 14 |
Hb_004326_010 |
0.1644362465 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_015675_030 |
0.1655153803 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002876_270 |
0.1660583932 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_021943_090 |
0.1660715956 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 18 |
Hb_000946_110 |
0.1674629815 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 19 |
Hb_001546_010 |
0.1680618735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010175_030 |
0.1704730919 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |