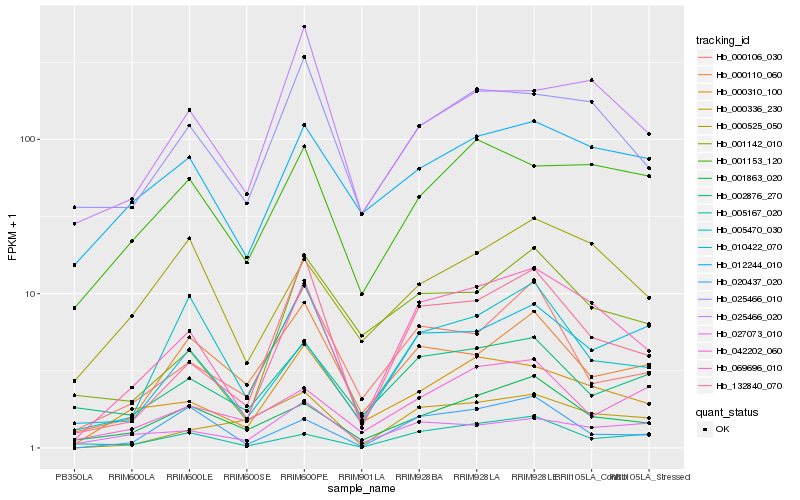
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_069696_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s21190g [Populus trichocarpa] |
| 2 |
Hb_132840_070 |
0.1464664887 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 3 |
Hb_000310_100 |
0.1555657574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_025466_010 |
0.1696618706 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001863_020 |
0.1729452287 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001142_010 |
0.1771535754 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |
| 7 |
Hb_010422_070 |
0.1783464949 |
- |
- |
PREDICTED: UPF0483 protein CBG03338-like [Jatropha curcas] |
| 8 |
Hb_001153_120 |
0.1822513442 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 9 |
Hb_012244_010 |
0.1833004328 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 10 |
Hb_000525_050 |
0.1856417267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005470_030 |
0.1857666552 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005167_020 |
0.1862753957 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 13 |
Hb_000336_230 |
0.1909225863 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa007609mg [Prunus persica] |
| 14 |
Hb_000106_030 |
0.1910866616 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_020437_020 |
0.1927990889 |
- |
- |
hypothetical protein POPTR_0001s01410g, partial [Populus trichocarpa] |
| 16 |
Hb_002876_270 |
0.1989094813 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000110_060 |
0.2038089231 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 18 |
Hb_027073_010 |
0.2045585951 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 19 |
Hb_025466_020 |
0.2069472347 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 20 |
Hb_042202_060 |
0.2102876589 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Jatropha curcas] |