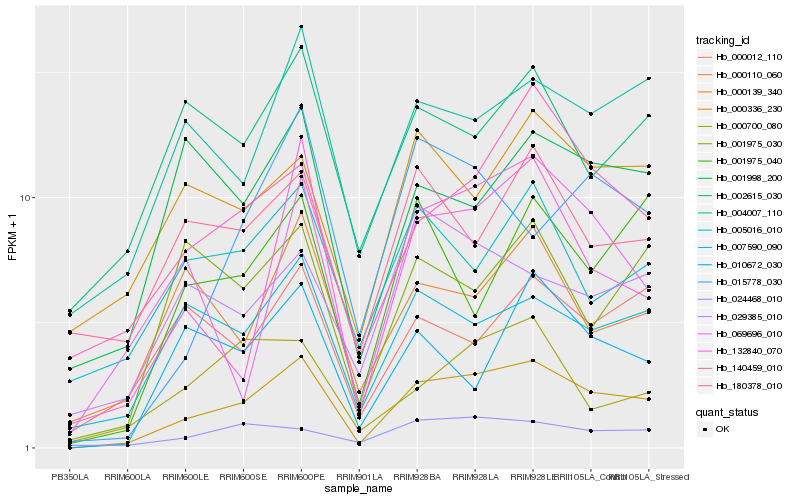
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_230 |
0.0 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa007609mg [Prunus persica] |
| 2 |
Hb_007590_090 |
0.1732714205 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 3 |
Hb_140459_010 |
0.177389081 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 4 |
Hb_132840_070 |
0.1789829313 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 5 |
Hb_004007_110 |
0.1811792384 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 6 |
Hb_024468_010 |
0.1842245722 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 7 |
Hb_069696_010 |
0.1909225863 |
- |
- |
hypothetical protein POPTR_0003s21190g [Populus trichocarpa] |
| 8 |
Hb_000110_060 |
0.1941620479 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 9 |
Hb_005016_010 |
0.1945526284 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 10 |
Hb_000139_340 |
0.1949152626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001998_200 |
0.19793649 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 12 |
Hb_000012_110 |
0.1982769143 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 13 |
Hb_001975_030 |
0.1987418039 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 14 |
Hb_010672_030 |
0.1997570244 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 15 |
Hb_000700_080 |
0.2000211986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001975_040 |
0.2001235287 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 17 |
Hb_180378_010 |
0.2003091484 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 18 |
Hb_002615_030 |
0.2016177041 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 19 |
Hb_015778_030 |
0.2035325039 |
- |
- |
PREDICTED: probable myosin light chain kinase DDB_G0279831 [Jatropha curcas] |
| 20 |
Hb_029385_010 |
0.2048920184 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |