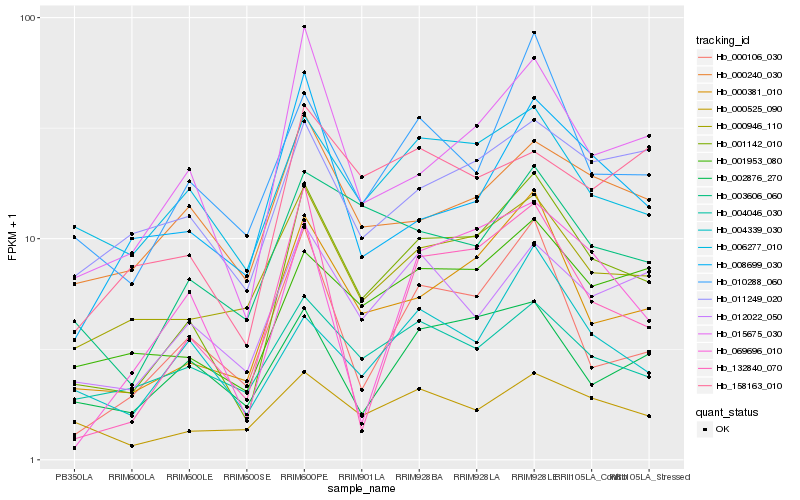
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001142_010 |
0.0 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |
| 2 |
Hb_000381_010 |
0.1084626654 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_015675_030 |
0.13423948 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001953_080 |
0.1443672155 |
- |
- |
pseudouridine synthase family protein, partial [Manihot esculenta] |
| 5 |
Hb_000106_030 |
0.1538078541 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_010288_060 |
0.1551996902 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 7 |
Hb_000946_110 |
0.1559502221 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 8 |
Hb_012022_050 |
0.1609938503 |
- |
- |
hypothetical protein EUGRSUZ_B02804 [Eucalyptus grandis] |
| 9 |
Hb_003606_060 |
0.1620830622 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 10 |
Hb_132840_070 |
0.1646051625 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 11 |
Hb_006277_010 |
0.1654270448 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 12 |
Hb_004339_030 |
0.1677468493 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 13 |
Hb_000525_090 |
0.1755062793 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 14 |
Hb_011249_020 |
0.1756219751 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 15 |
Hb_158163_010 |
0.1760083988 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_069696_010 |
0.1771535754 |
- |
- |
hypothetical protein POPTR_0003s21190g [Populus trichocarpa] |
| 17 |
Hb_004046_030 |
0.1786605553 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 18 |
Hb_008699_030 |
0.1787683787 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 19 |
Hb_002876_270 |
0.1790297712 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000240_030 |
0.179188398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |