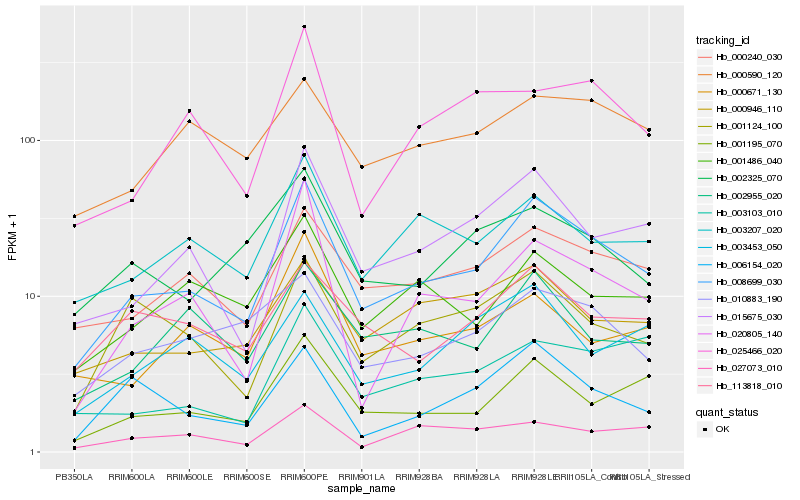
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008699_030 |
0.0 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 2 |
Hb_015675_030 |
0.1198921254 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000240_030 |
0.1278924279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001195_070 |
0.1316922941 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 5 |
Hb_003207_020 |
0.1427878972 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 6 |
Hb_000946_110 |
0.1519849448 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 7 |
Hb_001124_100 |
0.1536398601 |
- |
- |
PREDICTED: vesicle-associated protein 2-2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002955_020 |
0.1538105987 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_010883_190 |
0.1544040483 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006154_020 |
0.1552447134 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1621120 [Ricinus communis] |
| 11 |
Hb_001486_040 |
0.1558742099 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 12 |
Hb_020805_140 |
0.1594629125 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 13 |
Hb_113818_010 |
0.1594641671 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 14 |
Hb_003103_010 |
0.1606549314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002325_070 |
0.160681301 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_025466_020 |
0.1612451527 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 17 |
Hb_027073_010 |
0.1630937972 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 18 |
Hb_003453_050 |
0.1640958138 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 19 |
Hb_000590_120 |
0.1644184452 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 20 |
Hb_000671_130 |
0.1648709224 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |