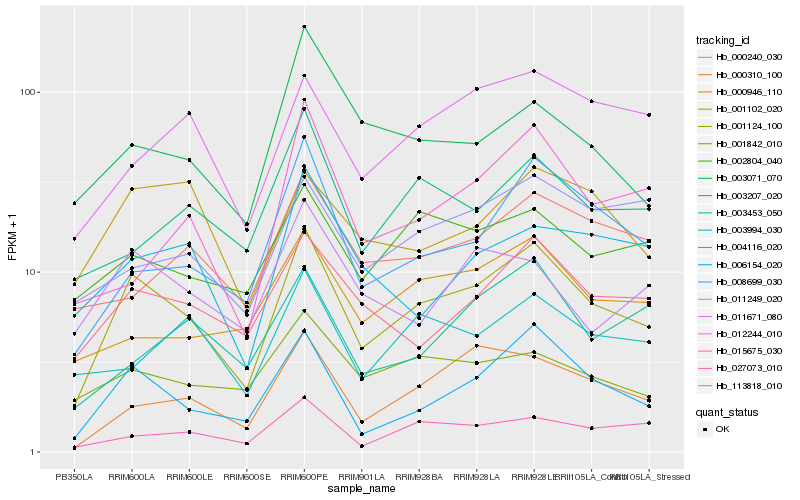
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001124_100 |
0.0 |
- |
- |
PREDICTED: vesicle-associated protein 2-2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_006154_020 |
0.1189044189 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1621120 [Ricinus communis] |
| 3 |
Hb_113818_010 |
0.1515036448 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 4 |
Hb_008699_030 |
0.1536398601 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 5 |
Hb_000310_100 |
0.1550829072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_027073_010 |
0.1612353035 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 7 |
Hb_002804_040 |
0.1669051642 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 8 |
Hb_012244_010 |
0.1695636875 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 9 |
Hb_000946_110 |
0.1695929071 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 10 |
Hb_001842_010 |
0.1704656715 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_001102_020 |
0.1718577927 |
- |
- |
solanesyl diphosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_003994_030 |
0.1746379503 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 13 |
Hb_003207_020 |
0.174886892 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 14 |
Hb_000240_030 |
0.1773734075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011249_020 |
0.1784127504 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 16 |
Hb_011671_080 |
0.1785668245 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 17 |
Hb_004116_020 |
0.1819101819 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 18 |
Hb_003453_050 |
0.1822149919 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 19 |
Hb_015675_030 |
0.1834601151 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003071_070 |
0.1835528267 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |