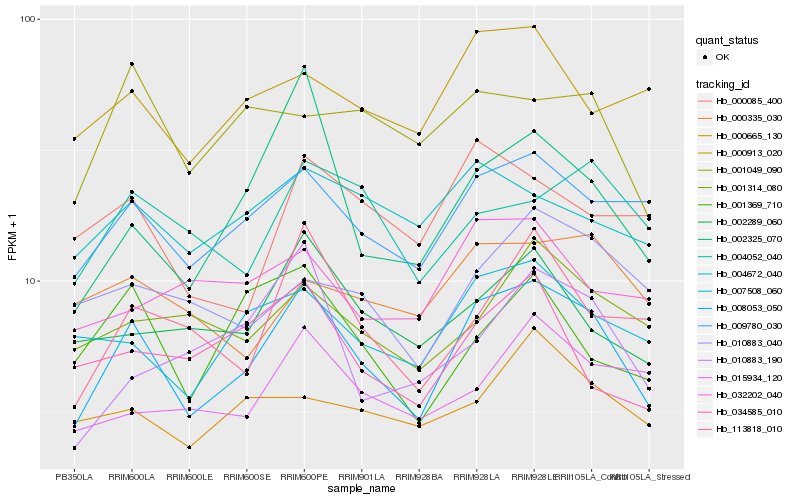
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008053_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 2 |
Hb_009780_030 |
0.1201768118 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 3 |
Hb_007508_060 |
0.1391946079 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 4 |
Hb_002325_070 |
0.1431558599 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_113818_010 |
0.1438111316 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 6 |
Hb_002289_060 |
0.1501090166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000913_020 |
0.1522117218 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 8 |
Hb_015934_120 |
0.1570368937 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 9 |
Hb_004672_040 |
0.1571093105 |
- |
- |
PREDICTED: SEC1 family transport protein SLY1-like [Populus euphratica] |
| 10 |
Hb_004052_040 |
0.1571146988 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 11 |
Hb_001369_710 |
0.1571727275 |
- |
- |
PREDICTED: uncharacterized protein LOC105648579 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001314_080 |
0.1577718275 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 13 |
Hb_000085_400 |
0.1582675258 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 14 |
Hb_001049_090 |
0.1599599784 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000665_130 |
0.1600808083 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 16 |
Hb_010883_040 |
0.1605538917 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 17 |
Hb_034585_010 |
0.1612661354 |
- |
- |
PREDICTED: uncharacterized protein LOC105639637 [Jatropha curcas] |
| 18 |
Hb_010883_190 |
0.1628480726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000335_030 |
0.1632092226 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17033 [Jatropha curcas] |
| 20 |
Hb_032202_040 |
0.1637968865 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |