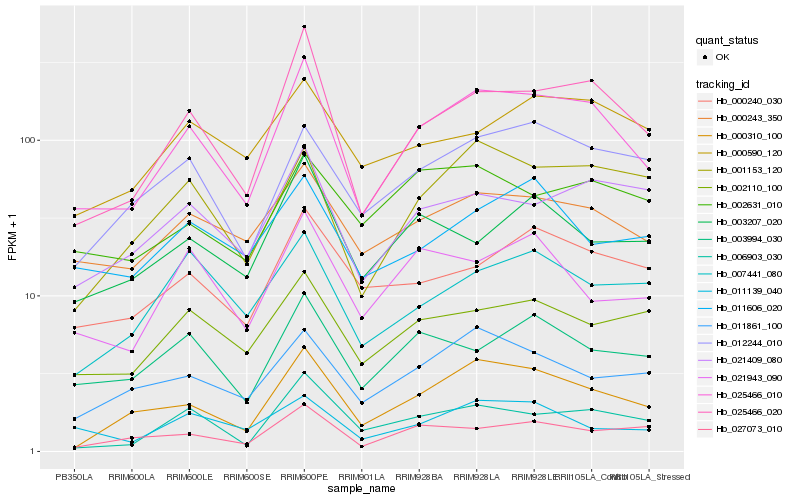
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025466_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_025466_020 |
0.088528257 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 3 |
Hb_000310_100 |
0.124238719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000243_350 |
0.1288333334 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 5 |
Hb_001153_120 |
0.1399878214 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 6 |
Hb_006903_030 |
0.1429182297 |
- |
- |
PREDICTED: uncharacterized protein LOC105640666 [Jatropha curcas] |
| 7 |
Hb_012244_010 |
0.143213711 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 8 |
Hb_003994_030 |
0.1446232678 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 9 |
Hb_021409_080 |
0.1446464492 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 10 |
Hb_000240_030 |
0.1465839872 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011861_100 |
0.1477132697 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_002110_100 |
0.1490810703 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_021943_090 |
0.1521133288 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 14 |
Hb_027073_010 |
0.1543385548 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 15 |
Hb_011139_040 |
0.1550200896 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_007441_080 |
0.1567717203 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 17 |
Hb_003207_020 |
0.1570137557 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 18 |
Hb_000590_120 |
0.1574487868 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 19 |
Hb_002631_010 |
0.162484164 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 20 |
Hb_011606_020 |
0.1631241516 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |