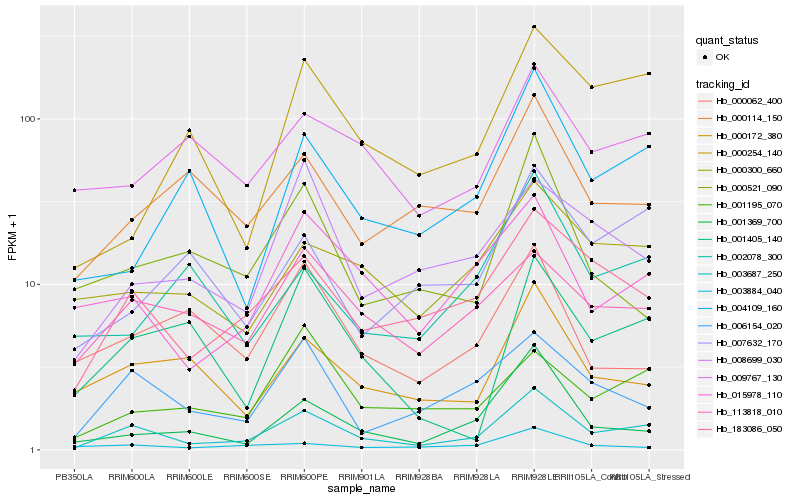
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003687_250 |
0.0 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 2 |
Hb_183086_050 |
0.169165548 |
- |
- |
PREDICTED: metal-nicotianamine transporter YSL3 [Jatropha curcas] |
| 3 |
Hb_006154_020 |
0.1911844572 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1621120 [Ricinus communis] |
| 4 |
Hb_015978_110 |
0.1950905059 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g62470, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_001369_700 |
0.2096673025 |
- |
- |
- |
| 6 |
Hb_000172_380 |
0.2104339712 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 7 |
Hb_113818_010 |
0.2107199554 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 8 |
Hb_000300_660 |
0.2156316825 |
- |
- |
unknown [Glycine max] |
| 9 |
Hb_001195_070 |
0.2199803658 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 10 |
Hb_004109_160 |
0.2205945336 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001405_140 |
0.2234337931 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 12 |
Hb_003884_040 |
0.2234424551 |
- |
- |
putative gag-pol polyprotein [Oryza sativa Japonica Group] |
| 13 |
Hb_008699_030 |
0.2258253866 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 14 |
Hb_000114_150 |
0.2264134909 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 15 |
Hb_009767_130 |
0.2294356664 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 16 |
Hb_000521_090 |
0.2315704065 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000254_140 |
0.2329734788 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic [Jatropha curcas] |
| 18 |
Hb_007632_170 |
0.2346956306 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 19 |
Hb_000062_400 |
0.2369497462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002078_300 |
0.2371204807 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |